Spatially Distinct Genetic Determinants of Aortic Dimensions Influence Risks of Aneurysm and Stenosis
- PMID: 35902171
- PMCID: PMC11216157
- DOI: 10.1016/j.jacc.2022.05.024
Spatially Distinct Genetic Determinants of Aortic Dimensions Influence Risks of Aneurysm and Stenosis
Abstract
Background: The left ventricular outflow tract (LVOT) and ascending aorta are spatially complex, with distinct pathologies and embryologic origins. Prior work examined the genetics of thoracic aortic diameter in a single plane.
Objectives: We sought to elucidate the genetic basis for the diameter of the LVOT, aortic root, and ascending aorta.
Methods: Using deep learning, we analyzed 2.3 million cardiac magnetic resonance images from 43,317 UK Biobank participants. We computed the diameters of the LVOT, the aortic root, and at 6 locations of ascending aorta. For each diameter, we conducted a genome-wide association study and generated a polygenic score. Finally, we investigated associations between these scores and disease incidence.
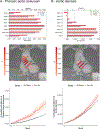
Results: A total of 79 loci were significantly associated with at least 1 diameter. Of these, 35 were novel, and most were associated with 1 or 2 diameters. A polygenic score of aortic diameter approximately 13 mm from the sinotubular junction most strongly predicted thoracic aortic aneurysm (n = 427,016; mean HR: 1.42 per SD; 95% CI: 1.34-1.50; P = 6.67 × 10-21). A polygenic score predicting a smaller aortic root was predictive of aortic stenosis (n = 426,502; mean HR: 1.08 per SD; 95% CI: 1.03-1.12; P = 5 × 10-6).
Conclusions: We detected distinct genetic loci underpinning the diameters of the LVOT, aortic root, and at several segments of ascending aorta. We spatially defined a region of aorta whose genetics may be most relevant to predicting thoracic aortic aneurysm. We further described a genetic signature that may predispose to aortic stenosis. Understanding genetic contributions to proximal aortic diameter may enable identification of individuals at risk for aortic disease and facilitate prioritization of therapeutic targets.
Keywords: aorta; cardiovascular disease; genetics; left ventricular outflow tract; machine learning.
Copyright © 2022 American College of Cardiology Foundation. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Funding Support and Author Disclosures This work was supported by the Fondation Leducq (14CVD01), and by grants from the National Institutes of Health (NIH) to Dr Pirruccello (K08HL159346), Dr Ellinor (1RO1HL092577, K24HL105780), and Dr Ho (R01HL134893, R01HL140224, K24HL153669). This work was also supported by a Sarnoff Cardiovascular Research Foundation Scholar Award to Dr Pirruccello. Dr Nauffal is supported by NIH grant 5T32HL007604-35. Dr Lubitz is supported by NIH grant R01HL139731 and American Heart Association 18SFRN34250007. This work was supported by a grant from the American Heart Association Strategically Focused Research Networks to Dr Ellinor. Dr Lindsay is supported by the Fredman Fellowship for Aortic Disease and the Toomey Fund for Aortic Dissection Research. This work was funded by a collaboration between the Broad Institute and IBM Research. Dr Pirruccello has served as a consultant for Maze Therapeutics. Dr Lubitz receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit; has consulted for Bristol Myers Squibb/Pfizer and Bayer AG; and participates in a research collaboration with IBM. Dr Ng is employed by IBM Research. Dr Batra receives sponsored research support from Bayer AG and IBM; and has consulted for Novartis and Prometheus Biosciences. Dr Ho has received research grant support from Bayer AG focused on machine learning and cardiovascular disease; and has received research supplies from EcoNugenics. Dr Philippakis is supported by a grant from Bayer AG to the Broad Institute focused on machine learning for clinical trial design. Dr Ellinor has received sponsored research support from Bayer AG and from IBM Research; and has served on advisory boards or consulted for Bayer AG, MyoKardia, and Novartis. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures



Comment in
-
Leveraging Machine Learning for Translational Genetics of Cardiovascular Imaging.J Am Coll Cardiol. 2022 Aug 2;80(5):498-499. doi: 10.1016/j.jacc.2022.05.020. J Am Coll Cardiol. 2022. PMID: 35902172 No abstract available.
References
-
- Grotenhuis HB, Dallaire F, Verpalen IM, van den Akker MJE, Mertens L, Friedberg MK. Aortic Root Dilatation and Aortic-Related Complications in Children After Tetralogy of Fallot Repair. Circ. Cardiovasc. Imaging 2018;11:e007611. - PubMed
-
- Axt-Fliedner R, Kreiselmaier P, Schwarze A, Krapp M, Gembruch U. Development of hypoplastic left heart syndrome after diagnosis of aortic stenosis in the first trimester by early echocardiography. Ultrasound Obstet. Gynecol 2006;28:106–109. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

