A modular spring-loaded actuator for mechanical activation of membrane proteins
- PMID: 35902570
- PMCID: PMC9334261
- DOI: 10.1038/s41467-022-30745-2
A modular spring-loaded actuator for mechanical activation of membrane proteins
Abstract
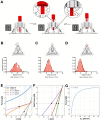
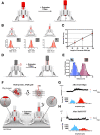
How cells respond to mechanical forces by converting them into biological signals underlie crucial cellular processes. Our understanding of mechanotransduction has been hindered by technical barriers, including limitations in our ability to effectively apply low range piconewton forces to specific mechanoreceptors on cell membranes without laborious and repetitive trials. To overcome these challenges we introduce the Nano-winch, a robust, easily assembled, programmable DNA origami-based molecular actuator. The Nano-winch is designed to manipulate multiple mechanoreceptors in parallel by exerting fine-tuned, low- piconewton forces in autonomous and remotely activated modes via adjustable single- and double-stranded DNA linkages, respectively. Nano-winches in autonomous mode can land and operate on the cell surface. Targeting the device to integrin stimulated detectable downstream phosphorylation of focal adhesion kinase, an indication that Nano-winches can be applied to study cellular mechanical processes. Remote activation mode allowed finer extension control and greater force exertion. We united remotely activated Nano-winches with single-channel bilayer experiments to directly observe the opening of a channel by mechanical force in the force responsive gated channel protein, BtuB. This customizable origami provides an instrument-free approach that can be applied to control and explore a diversity of mechanotransduction circuits on living cells.
© 2022. The Author(s).
Conflict of interest statement
INSERM (Institut National de la Santé et de la Recherche Médicale), Centre national de la recherche scientifique, and Université de Montpellier have submitted a patent application to the European patent Office pertaining to a nucleic acid origami modular device which can be used for exerting high throughput mechanical forces and constraints on surfaces (application number EP22305969.2). G.B. and A.M. are co-inventors, the remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

