The Type III Secretion Effector CteG Mediates Host Cell Lytic Exit of Chlamydia trachomatis
- PMID: 35903198
- PMCID: PMC9318579
- DOI: 10.3389/fcimb.2022.902210
The Type III Secretion Effector CteG Mediates Host Cell Lytic Exit of Chlamydia trachomatis
Abstract
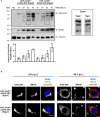
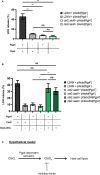
Chlamydia trachomatis is an obligate intracellular bacterium causing ocular and urogenital infections in humans that are a significant burden worldwide. The completion of its characteristic infectious cycle relies on the manipulation of several host cell processes by numerous chlamydial type III secretion effector proteins. We previously identified the C. trachomatis CteG effector and showed it localizes at the host cell plasma membrane at late stages of infection. Here, we showed that, from 48 h post-infection, mammalian cells infected by wild-type C. trachomatis contained more infectious chlamydiae in the culture supernatant than cells infected by a CteG-deficient strain. This phenotype was CteG-dependent as it could be complemented in cells infected by the CteG-deficient strain carrying a plasmid encoding CteG. Furthermore, we detected a CteG-dependent defect on host cell cytotoxicity, indicating that CteG mediates chlamydial lytic exit. Previous studies showed that Pgp4, a global regulator of transcription encoded in the C. trachomatis virulence plasmid, also mediates chlamydial lytic exit. However, by using C. trachomatis strains encoding or lacking Pgp4, we showed that production and localization of CteG are not regulated by Pgp4. A C. trachomatis strain lacking both CteG and Pgp4 was as defective in promoting host cell cytotoxicity as mutant strains lacking only CteG or Pgp4. Furthermore, CteG overproduction in a plasmid suppressed the host cell cytotoxic defect of CteG- and Pgp4-deficient chlamydiae. Overall, we revealed the first chlamydial type III secretion effector involved in host cell lytic exit. Our data indicates that CteG and Pgp4 participate in a single cascade of events, but involving multiple layers of regulation, leading to lysis of host cells and release of the infectious chlamydiae.
Keywords: Chlamydia trachomatis; effectors; host-pathogen interactions; pathogen egress; type III secretion.
Copyright © 2022 Pereira, Pais, Borges, Borrego, Gomes and Mota.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of homologs of the Chlamydia trachomatis effector CteG reveals a family of Chlamydiaceae type III secreted proteins that can be delivered into host cells.Med Microbiol Immunol. 2024 Jul 15;213(1):15. doi: 10.1007/s00430-024-00798-9. Med Microbiol Immunol. 2024. PMID: 39008129 Free PMC article.
-
Chlamydial Lytic Exit from Host Cells Is Plasmid Regulated.mBio. 2015 Nov 10;6(6):e01648-15. doi: 10.1128/mBio.01648-15. mBio. 2015. PMID: 26556273 Free PMC article.
-
CteG is a Chlamydia trachomatis effector protein that associates with the Golgi complex of infected host cells.Sci Rep. 2019 Apr 16;9(1):6133. doi: 10.1038/s41598-019-42647-3. Sci Rep. 2019. PMID: 30992493 Free PMC article.
-
Chlamydial Plasmid-Dependent Pathogenicity.Trends Microbiol. 2017 Feb;25(2):141-152. doi: 10.1016/j.tim.2016.09.006. Epub 2016 Oct 3. Trends Microbiol. 2017. PMID: 27712952 Free PMC article. Review.
-
Got mutants? How advances in chlamydial genetics have furthered the study of effector proteins.Pathog Dis. 2021 Feb 4;79(2):ftaa078. doi: 10.1093/femspd/ftaa078. Pathog Dis. 2021. PMID: 33512479 Free PMC article. Review.
Cited by
-
In Search of a Mechanistic Link between Chlamydia trachomatis-Induced Cellular Pathophysiology and Oncogenesis.Infect Immun. 2023 Feb 16;91(2):e0044322. doi: 10.1128/iai.00443-22. Epub 2023 Jan 25. Infect Immun. 2023. PMID: 36695575 Free PMC article. Review.
-
Advances in genetic manipulation of Chlamydia trachomatis.Front Immunol. 2023 Jun 28;14:1209879. doi: 10.3389/fimmu.2023.1209879. eCollection 2023. Front Immunol. 2023. PMID: 37449211 Free PMC article. Review.
-
TargeTron inactivation of plasmid-regulated Chlamydia trachomatis CT084 results in a nonlytic phenotype.Pathog Dis. 2023 Jan 17;81:ftad026. doi: 10.1093/femspd/ftad026. Pathog Dis. 2023. PMID: 37804183 Free PMC article.
-
Plasmid-mediated virulence in Chlamydia.Front Cell Infect Microbiol. 2023 Aug 17;13:1251135. doi: 10.3389/fcimb.2023.1251135. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37662000 Free PMC article. Review.
-
The Chlamydia trachomatis IncM Protein Interferes with Host Cell Cytokinesis, Centrosome Positioning, and Golgi Distribution and Contributes to the Stability of the Pathogen-Containing Vacuole.Infect Immun. 2023 Apr 18;91(4):e0040522. doi: 10.1128/iai.00405-22. Epub 2023 Mar 6. Infect Immun. 2023. PMID: 36877064 Free PMC article.
References
-
- Batteiger B. (2012). “Chlamydia Infection and Epidemiology,” in Intracellular Pathogens I ,” in Chlamydiales. Eds. Tan M., Bavoil P. (Washington, DC: ASM Press; ), 1–26. doi: 10.1128/9781555817329 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

