Finding Correlations Between mRNA and Protein Levels in Leishmania Development: Is There a Discrepancy?
- PMID: 35903202
- PMCID: PMC9318571
- DOI: 10.3389/fcimb.2022.852902
Finding Correlations Between mRNA and Protein Levels in Leishmania Development: Is There a Discrepancy?
Abstract
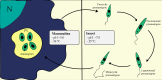
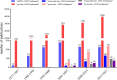
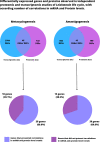
Multiple genes and proteins have been identified as differentially expressed in the stages of the Leishmania life cycle. The differentiation processes are implicated in specific transcriptional and proteomic adjustments driven by gene expression regulation mechanisms. Leishmania parasites lack gene-specific transcriptional control, and gene expression regulation mostly depends on posttranscriptional mechanisms. Due to the lack of transcriptional regulation, criticism regarding the relevance of transcript quantification as a possible and efficient prediction of protein levels is recurrent in studies that use transcriptomic information. The advent of high-throughput technologies has improved the analysis of genomes, transcriptomes and proteomes for different organisms under several conditions. Nevertheless, defining the correlation between transcriptional and proteomic profiles requires arduous and expensive work and remains a challenge in Leishmania. In this review, we analyze transcriptomic and proteomic data for several Leishmania species in two different stages of the parasite life cycle: metacyclogenesis and amastigogenesis (amastigote differentiation). We found a correlation between mRNA and protein levels of 60.9% and 69.8% for metacyclogenesis and amastigogenesis, respectively; showing that majority mRNA and protein levels increase or decrease concomitantly. Among the analyzed genes that did not present correlation indicate that transcriptomic data should be carefully interpreted as protein expression. We also discuss possible explanations and mechanisms involved for this lack of correlation.
Keywords: amastigote differentiation; gene expression; life cycle; metacyclogenesis; proteome; transcriptome.
Copyright © 2022 Cortazzo da Silva, Aoki and Floeter-Winter.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor LT declared a shared affiliation with the authors at the time of review.
Figures

Similar articles
-
The mRNA-bound Proteome of Leishmania mexicana: Novel Genetic Insight into an Ancient Parasite.Mol Cell Proteomics. 2019 Jul;18(7):1271-1284. doi: 10.1074/mcp.RA118.001307. Epub 2019 Apr 4. Mol Cell Proteomics. 2019. PMID: 30948621 Free PMC article.
-
A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum.Proteomics. 2006 Jun;6(12):3567-81. doi: 10.1002/pmic.200500853. Proteomics. 2006. PMID: 16705753
-
Developmental differentiation in Leishmania lifecycle progression: post-transcriptional control conducts the orchestra.Curr Opin Microbiol. 2016 Dec;34:82-89. doi: 10.1016/j.mib.2016.08.004. Epub 2016 Sep 9. Curr Opin Microbiol. 2016. PMID: 27565628 Review.
-
Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed.Mol Biochem Parasitol. 2007 Mar;152(1):35-46. doi: 10.1016/j.molbiopara.2006.11.009. Epub 2006 Dec 8. Mol Biochem Parasitol. 2007. PMID: 17188763
-
Global gene expression in Leishmania.Int J Parasitol. 2007 Aug;37(10):1077-86. doi: 10.1016/j.ijpara.2007.04.011. Epub 2007 May 6. Int J Parasitol. 2007. PMID: 17574557 Review.
Cited by
-
The Elusive Trypanosoma cruzi Disperse Gene Protein Family (DGF-1).Pathogens. 2023 Feb 10;12(2):292. doi: 10.3390/pathogens12020292. Pathogens. 2023. PMID: 36839564 Free PMC article. Review.
-
Schisandra extract ameliorates arthritis pathogenesis by suppressing the NF-κB and MAPK signalling pathways.J Cell Mol Med. 2023 Jul;27(14):2071-2081. doi: 10.1111/jcmm.17814. Epub 2023 Jun 20. J Cell Mol Med. 2023. PMID: 37337779 Free PMC article.
-
Expression of SOX10, and micro-RNA 221 and Micro-RNA 222 in Oral Squamous Cell Carcinoma and Correlation with Clinicopathological Characteristics.Head Neck Pathol. 2025 Jun 25;19(1):76. doi: 10.1007/s12105-025-01809-8. Head Neck Pathol. 2025. PMID: 40560444
-
Proteomic data of the Trypanosoma cruzi insect-dwelling epimastigotes overexpressing the RNA-binding protein UBP1.Data Brief. 2024 Jan 24;53:110085. doi: 10.1016/j.dib.2024.110085. eCollection 2024 Apr. Data Brief. 2024. PMID: 38348324 Free PMC article.
-
Molecular Mechanisms of Drug Resistance in Leishmania spp.Pathogens. 2024 Sep 27;13(10):835. doi: 10.3390/pathogens13100835. Pathogens. 2024. PMID: 39452707 Free PMC article. Review.
References
-
- Alcolea P. J., Alonso A., Gómez M. J., Sánchez-Gorostiaga A., Moreno-Paz M., González-Pastor E., et al. . (2010. b). Temperature Increase Prevails Over Acidification in Gene Expression Modulation of Amastigote Differentiation in Leishmania Infantum. BMC Genomics 11:1–24. doi: 10.1186/1471-2164-11-31 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

