Genetic Diversity and Selection Signatures in Synthetic-Derived Wheats and Modern Spring Wheat
- PMID: 35903232
- PMCID: PMC9315363
- DOI: 10.3389/fpls.2022.877496
Genetic Diversity and Selection Signatures in Synthetic-Derived Wheats and Modern Spring Wheat
Abstract
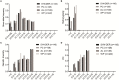
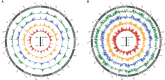
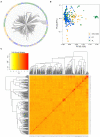
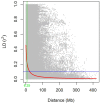
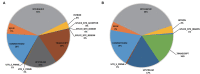
Synthetic hexaploid wheats and their derived advanced lines were subject to empirical selection in developing genetically superior cultivars. To investigate genetic diversity, patterns of nucleotide diversity, population structure, and selection signatures during wheat breeding, we tested 422 wheat accessions, including 145 synthetic-derived wheats, 128 spring wheat cultivars, and 149 advanced breeding lines from Pakistan. A total of 18,589 high-quality GBS-SNPs were identified that were distributed across the A (40%), B (49%), and D (11%) genomes. Values of population diversity parameters were estimated across chromosomes and genomes. Genome-wide average values of genetic diversity and polymorphic information content were estimated to be 0.30 and 0.25, respectively. Neighbor-joining (NJ) tree, principal component analysis (PCA), and kinship analyses revealed that synthetic-derived wheats and advanced breeding lines were genetically diverse. The 422 accessions were not separated into distinct groups by NJ analysis and confirmed using the PCA. This conclusion was validated with both relative kinship and Rogers' genetic distance analyses. EigenGWAS analysis revealed that 32 unique genome regions had undergone selection. We found that 50% of the selected regions were located in the B-genome, 29% in the D-genome, and 21% in the A-genome. Previously known functional genes or QTL were found within the selection regions associated with phenology-related traits such as vernalization, adaptability, disease resistance, and yield-related traits. The selection signatures identified in the present investigation will be useful for understanding the targets of modern wheat breeding in Pakistan.
Keywords: EigenGWAS; bread wheat; gene annotation; genetic diversity; genotyping-by-sequencing; selection signatures.
Copyright © 2022 Ali, Danting, Wang, Sadiq, Rasheed, He and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Afzal F., Reddy B., Gul A., Khalid M., Subhani A., Shazadi K. (2017). Physiological, biochemical and agronomic traits associated with drought tolerance in a synthetic-derived wheat diversity panel. Crop Pasture Sci. 68, 213–224. 10.1071/CP16367 - DOI
LinkOut - more resources
Full Text Sources

