Spin-Labeled Riboswitch Synthesized from a Protected TPA Phosphoramidite Building Block
- PMID: 35903916
- PMCID: PMC9804336
- DOI: 10.1002/chem.202201822
Spin-Labeled Riboswitch Synthesized from a Protected TPA Phosphoramidite Building Block
Abstract
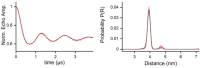
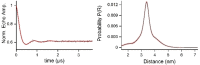
The nitroxide TPA (2,2,5,5-tetramethyl-pyrrolin-1-oxyl-3-acetylene) is an excellent spin label for EPR studies of RNA. Previous synthetic methods, however, are complicated and require special equipment. Herein, we describe a uridine derived phosphoramidite with a photocaged TPA unit attached. The light sensitive 2-nitrobenzyloxymethyl group can be removed in high yield by short irradiation at 365 nm. Based on this approach, a doubly spin-labeled 27mer neomycin sensing riboswitch was synthesized and studied by PELDOR. The overall thermal stability of the fold is not much reduced by TPA. In-line probing nevertheless detected changes in local mobility.
Keywords: PELDOR; RNA aptamer; nitroxide; photocaged spin label; secondary structure mapping.
© 2022 The Authors. Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Torricella F., Pierro A., Mileo E., Belle V., Bonucci A., Biochim. Biophys. Acta 2021, 1869, 140653. - PubMed
-
- Haugland M. M., Lovett J. E., Anderson E. A., Chem. Soc. Rev. 2018, 47, 668–680. - PubMed
-
- Endeward B., Marko A., Denysenkov V. P., Sigurdsson S. T., Prisner T. F., Methods Enzymol. 2015, 564, 403–425. - PubMed
-
- Stelzl L. S., Erlenbach N., Heinz M., Prisner T. F., Hummer G. J., J. Am. Chem. Soc. 2017, 139, 11674–11677. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

