Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis
- PMID: 35904863
- PMCID: PMC9558494
- DOI: 10.1002/anie.202203560
Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis
Abstract
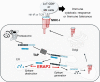
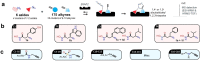
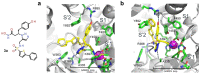
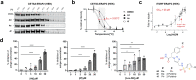
Endoplasmic reticulum aminopeptidase 2 (ERAP2) is a key enzyme involved in the trimming of antigenic peptides presented by Major Histocompatibility Complex class I. It is a target of growing interest for the treatment of autoimmune diseases and in cancer immunotherapy. However, the discovery of potent and selective ERAP2 inhibitors is highly challenging. Herein, we have used kinetic target-guided synthesis (KTGS) to identify such inhibitors. Co-crystallization experiments revealed the binding mode of three different inhibitors with increasing potency and selectivity over related enzymes. Selected analogues engage ERAP2 in cells and inhibit antigen presentation in a cellular context. 4 d (BDM88951) displays favorable in vitro ADME properties and in vivo exposure. In summary, KTGS allowed the discovery of the first nanomolar and selective highly promising ERAP2 inhibitors that pave the way of the exploration of the biological roles of this enzyme and provide lead compounds for drug discovery efforts.
Keywords: ERAP2; Isoform Selectivity; Medicinal Chemistry; Metalloenzymes; Protein-Templated Reactions.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Evnouchidou I., van Endert P., Hum. Immunol. 2019, 80, 290–295. - PubMed
-
- Yao Y., Liu N., Zhou Z., Shi L., Hum. Immunol. 2019, 80, 325–334. - PubMed
-
- Compagnone M., Cifaldi L., Fruci D., Hum. Immunol. 2019, 80, 318–324. - PubMed
-
- de Castro J. A. L., Stratikos E., Hum. Immunol. 2019, 80, 310–317. - PubMed

