Helicobacter pylori actively suppresses innate immune nucleic acid receptors
- PMID: 35905376
- PMCID: PMC9341374
- DOI: 10.1080/19490976.2022.2105102
Helicobacter pylori actively suppresses innate immune nucleic acid receptors
Abstract
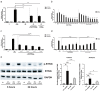
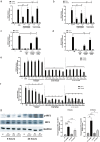
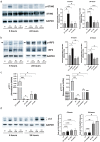
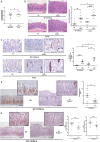
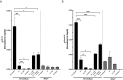
Chronic mucosal pathogens have evolved multiple strategies to manipulate the host immune response; consequently, microbes contribute to the development of >2 million cases of cancer/year. Gastric adenocarcinoma is the fourth leading cause of cancer-related death and Helicobacter pylori confers the highest risk for this disease. Gastric innate immune effectors can either eliminate bacteria or mobilize adaptive immune responses including Toll-like receptors (TLRs), and cytosolic DNA sensor/adaptor proteins (e.g., stimulator of interferon genes, STING). The H. pylori strain-specific cag type IV secretion system (T4SS) augments gastric cancer risk and translocates DNA into epithelial cells where it activates the microbial DNA sensor TLR9 and suppresses injury in vivo; however, the ability of H. pylori to suppress additional nucleic acid PRRs within the context of chronic gastric inflammation and injury remains undefined. In this study, in vitro and ex vivo experiments identified a novel mechanism through which H. pylori actively suppresses STING and RIG-I signaling via downregulation of IRF3 activation. In vivo, the use of genetically deficient mice revealed that Th17 inflammatory responses are heightened following H. pylori infection within the context of Sting deficiency in conjunction with increased expression of a known host immune regulator, Trim30a. This novel mechanism of immune suppression by H. pylori is likely a critical component of a finely tuned rheostat that not only regulates the initial innate immune response, but also drives chronic gastric inflammation and injury.
Keywords: Helicobacter pylori; RIG-I; STING; TLR9; TRIM30; gastric cancer; innate immunity.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
