Asymmetric Histone Inheritance: Establishment, Recognition, and Execution
- PMID: 35905975
- PMCID: PMC10054593
- DOI: 10.1146/annurev-genet-072920-125226
Asymmetric Histone Inheritance: Establishment, Recognition, and Execution
Abstract
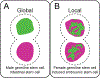
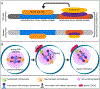
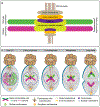
The discovery of biased histone inheritance in asymmetrically dividing Drosophila melanogaster male germline stem cells demonstrates one means to produce two distinct daughter cells with identical genetic material. This inspired further studies in different systems, which revealed that this phenomenon may be a widespread mechanism to introduce cellular diversity. While the extent of asymmetric histone inheritance could vary among systems, this phenomenon is proposed to occur in three steps: first, establishment of histone asymmetry between sister chromatids during DNA replication; second, recognition of sister chromatids carrying asymmetric histone information during mitosis; and third, execution of this asymmetry in the resulting daughter cells. By compiling the current knowledge from diverse eukaryotic systems, this review comprehensively details and compares known chromatin factors, mitotic machinery components, and cell cycle regulators that may contribute to each of these three steps. Also discussed are potential mechanisms that introduce and regulate variable histone inheritance modes and how these different modes may contribute to cell fate decisions in multicellular organisms.
Keywords: DNA replication–coupled histone assembly; epigenetic inheritance; epigenetic memory; mitotic drive; nonrandom chromatid segregation; nucleosome density.
Conflict of interest statement
Figures



References
Literature cited:
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

