Genomic annotation and molecular evolution of monkeypox virus outbreak in 2022
- PMID: 35906185
- PMCID: PMC10087776
- DOI: 10.1002/jmv.28036
Genomic annotation and molecular evolution of monkeypox virus outbreak in 2022
Abstract
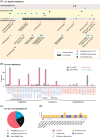
Monkeypox virus (MPXV) has generally circulated in West and Central Africa since its emergence. Recently, sporadic MPXV infections in several nonendemic countries have attracted widespread attention. Here, we conducted a systematic analysis of the recent outbreak of MPXV-2022, including its genomic annotation and molecular evolution. The phylogenetic analysis indicated that the MPXV-2022 strains belong to the same lineage of the MPXV strain isolated in 2018. However, compared with the MPXV strain in 2018, in total 46 new consensus mutations were observed in the MPXV-2022 strains, including 24 nonsynonymous mutations. By assigning mutations to 187 proteins encoded by the MPXV genome, we found that 10 proteins in the MPXV are more prone to mutation, including D2L-like, OPG023, OPG047, OPG071, OPG105, OPG109, A27L-like, OPG153, OPG188, and OPG210 proteins. In the MPXV-2022 strains, four and three nucleotide substitutions are observed in OPG105 and OPG210, respectively. Overall, our studies illustrated the genome evolution of the ongoing MPXV outbreak and pointed out novel mutations as a reference for further studies.
Keywords: MPXV endemic; molecular evolution; monkeypox; orthopoxvirus.
© 2022 The Authors. Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Marennikova S, Gurvich E, Shelukhina E. Comparison of the properties of five pox virus strains isolated from monkeys. Arch Gesamte Virusforsch. 1971;33(3):201‐210. - PubMed
-
- Control CfD, Prevention . Multistate outbreak of monkeypox—Illinois, Indiana, and Wisconsin, 2003. MMWR Morb Mort Wkly Rep. 2003;52(23):537‐540. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

