Functional characterization of FBXL7 as a novel player in human cancers
- PMID: 35906197
- PMCID: PMC9338262
- DOI: 10.1038/s41420-022-01143-w
Functional characterization of FBXL7 as a novel player in human cancers
Abstract
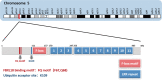
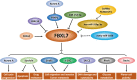
F-box and leucine-rich repeat protein 7 (FBXL7), an F-box protein responsible for substrate recognition by the SKP1-Cullin-1-F-box (SCF) ubiquitin ligases, plays an emerging role in the regulation of tumorigenesis and tumor progression. FBXL7 promotes polyubiquitylation and degradation of diverse substrates and is involved in many biological processes, including apoptosis, cell proliferation, cell migration and invasion, tumor metastasis, DNA damage, glucose metabolism, planar cell polarity, and drug resistance. In this review, we summarize the downstream substrates and upstream regulators of FBXL7. We then discuss its role in tumorigenesis and tumor progression as either an oncoprotein or a tumor suppressor, and further describe its aberrant expression and association with patient survival in human cancers. Finally, we provide future perspectives on validating FBXL7 as a cancer biomarker for diagnosis and prognosis and/or as a potential therapeutic target for anticancer treatment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
F-Box and Leucine-Rich Repeat Protein 7 Is a Prognostic Biomarker and Is Correlated with the Immunosuppressive Microenvironment in Colorectal Cancer.Genet Test Mol Biomarkers. 2023 Oct;27(10):325-338. doi: 10.1089/gtmb.2023.0075. Epub 2023 Oct 20. Genet Test Mol Biomarkers. 2023. PMID: 37862037
-
F-box protein Fbxl18 mediates polyubiquitylation and proteasomal degradation of the pro-apoptotic SCF subunit Fbxl7.Cell Death Dis. 2015 Feb 5;6(2):e1630. doi: 10.1038/cddis.2014.585. Cell Death Dis. 2015. PMID: 25654763 Free PMC article.
-
The Proapoptotic F-box Protein Fbxl7 Regulates Mitochondrial Function by Mediating the Ubiquitylation and Proteasomal Degradation of Survivin.J Biol Chem. 2015 May 8;290(19):11843-52. doi: 10.1074/jbc.M114.629931. Epub 2015 Mar 16. J Biol Chem. 2015. PMID: 25778398 Free PMC article.
-
The characteristics and roles of β-TrCP1/2 in carcinogenesis.FEBS J. 2021 Jun;288(11):3351-3374. doi: 10.1111/febs.15585. Epub 2020 Oct 23. FEBS J. 2021. PMID: 33021036 Review.
-
Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases.Cell Res. 2013 May;23(5):599-619. doi: 10.1038/cr.2013.44. Epub 2013 Mar 26. Cell Res. 2013. PMID: 23528706 Free PMC article. Review.
Cited by
-
Establishment of a circRNA-regulated E3 ubiquitin ligase signature and nomogram to predict immunotherapeutic efficacy and prognosis in hepatocellular carcinoma.Eur J Med Res. 2024 Jun 10;29(1):318. doi: 10.1186/s40001-024-01893-6. Eur J Med Res. 2024. PMID: 38858746 Free PMC article.
-
Genome-wide DNA methylation profiling of blood samples from patients with major depressive disorder: correlation with symptom heterogeneity.J Psychiatry Neurosci. 2025 Apr 2;50(2):E112-E124. doi: 10.1503/jpn.240126. Print 2025 Mar-Apr. J Psychiatry Neurosci. 2025. PMID: 40174930 Free PMC article.
-
Genome-wide 5-hydroxymethylcytosines in circulating cell-free DNA as noninvasive diagnostic markers for gastric cancer.Gastric Cancer. 2024 Jul;27(4):735-746. doi: 10.1007/s10120-024-01493-7. Epub 2024 Apr 7. Gastric Cancer. 2024. PMID: 38584223
References
Publication types
Grants and funding
- 92053117/National Natural Science Foundation of China (National Science Foundation of China)
- 81972591/National Natural Science Foundation of China (National Science Foundation of China)
- 82188102/National Natural Science Foundation of China (National Science Foundation of China)
- 81974429/National Natural Science Foundation of China (National Science Foundation of China)
- 82172898/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources

