HSF1 phosphorylation establishes an active chromatin state via the TRRAP-TIP60 complex and promotes tumorigenesis
- PMID: 35906200
- PMCID: PMC9338313
- DOI: 10.1038/s41467-022-32034-4
HSF1 phosphorylation establishes an active chromatin state via the TRRAP-TIP60 complex and promotes tumorigenesis
Abstract
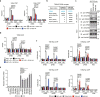
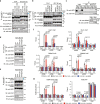
Transcriptional regulation by RNA polymerase II is associated with changes in chromatin structure. Activated and promoter-bound heat shock transcription factor 1 (HSF1) recruits transcriptional co-activators, including histone-modifying enzymes; however, the mechanisms underlying chromatin opening remain unclear. Here, we demonstrate that HSF1 recruits the TRRAP-TIP60 acetyltransferase complex in HSP72 promoter during heat shock in a manner dependent on phosphorylation of HSF1-S419. TRIM33, a bromodomain-containing ubiquitin ligase, is then recruited to the promoter by interactions with HSF1 and a TIP60-mediated acetylation mark, and cooperates with the related factor TRIM24 for mono-ubiquitination of histone H2B on K120. These changes in histone modifications are triggered by phosphorylation of HSF1-S419 via PLK1, and stabilize the HSF1-transcription complex in HSP72 promoter. Furthermore, HSF1-S419 phosphorylation is constitutively enhanced in and promotes proliferation of melanoma cells. Our results provide mechanisms for HSF1 phosphorylation-dependent establishment of an active chromatin status, which is important for tumorigenesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

