Using HPV-meta for human papillomavirus RNA quality detection
- PMID: 35906372
- PMCID: PMC9338075
- DOI: 10.1038/s41598-022-17318-5
Using HPV-meta for human papillomavirus RNA quality detection
Abstract
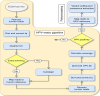
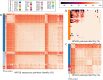
In the era of cervical cancer elimination, accurate and validated pipelines to detect human papillomavirus are essential to elucidate and understand HPV association with human cancers. We aimed to provide an open-source pipeline, "HPV-meta", to detect HPV transcripts in RNA sequencing data, including several steps to warn operators for possible viral contamination. The "HPV-meta" pipeline automatically performs several steps, starting with quality trimming, human genome filtering, HPV detection (blastx), cut-off settlement (10 reads and 690 bp coverage to make an HPV call) and finishing with fasta sequence generation for HPV positive samples. Fasta sequences can then be aligned to assess sequence diversity among HPV positive samples. All RNA sequencing files (n = 10,908) present in the cancer genome atlas (TCGA) were analyzed. "HPV-meta" identified 25 different HPV types being present in 488/10,904 specimens. Validation of results showed 99.98% agreement (10,902/10,904). Multiple alignment from fasta files warned about high sequence identity between several HPV 18 and 38 positive samples, whose contamination had previously been reported. The "HPV-meta" pipeline is a robust and validated pipeline that detects HPV in RNA sequencing data. Obtaining the fasta files enables contamination investigation, a non very rare occurrence in next generation sequencing.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Human papillomaviruses. IARC Monographs on the Evaluation of Carcinogenic

