Microbiome of the freshwater sponge Ephydatia muelleri shares compositional and functional similarities with those of marine sponges
- PMID: 35906397
- PMCID: PMC9562138
- DOI: 10.1038/s41396-022-01296-7
Microbiome of the freshwater sponge Ephydatia muelleri shares compositional and functional similarities with those of marine sponges
Abstract
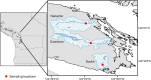
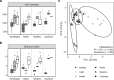
Sponges are known for hosting diverse communities of microbial symbionts, but despite persistent interest in the sponge microbiome, most research has targeted marine sponges; freshwater sponges have been the focus of less than a dozen studies. Here, we used 16 S rRNA gene amplicon sequencing and shotgun metagenomics to characterize the microbiome of the freshwater sponge Ephydatia muelleri and identify potential indicators of sponge-microbe mutualism. Using samples collected from the Sooke, Nanaimo, and Cowichan Rivers on Vancouver Island, British Columbia, we show that the E. muelleri microbiome is distinct from the ambient water and adjacent biofilms and is dominated by Sediminibacterium, Comamonas, and unclassified Rhodospirillales. We also observed phylotype-level differences in sponge microbiome taxonomic composition among different rivers. These differences were not reflected in the ambient water, suggesting that other environmental or host-specific factors may drive the observed geographic variation. Shotgun metagenomes and metagenome-assembled genomes further revealed that freshwater sponge-associated bacteria share many genomic similarities with marine sponge microbiota, including an abundance of defense-related proteins (CRISPR, restriction-modification systems, and transposases) and genes for vitamin B12 production. Overall, our results provide foundational information on the composition and function of freshwater sponge-associated microbes, which represent an important yet underappreciated component of the global sponge microbiome.
© 2022. The Author(s), under exclusive licence to International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- van Soest RWM, Boury-Esnault N, Hooper JNA, Rützler K, de Voogd NJ, Alvarez B, et al. World Porifera Database. http://www.marinespecies.org/porifera.

