Iron oxide and iron oxyhydroxide nanoparticles impair SARS-CoV-2 infection of cultured cells
- PMID: 35907835
- PMCID: PMC9338509
- DOI: 10.1186/s12951-022-01542-2
Iron oxide and iron oxyhydroxide nanoparticles impair SARS-CoV-2 infection of cultured cells
Abstract
Background: Coronaviruses usually cause mild respiratory disease in humans but as seen recently, some human coronaviruses can cause more severe diseases, such as the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the global spread of which has resulted in the ongoing coronavirus pandemic.
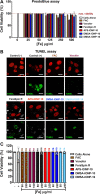
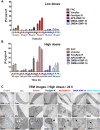
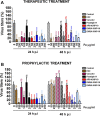
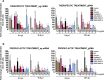
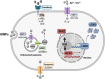
Results: In this study we analyzed the potential of using iron oxide nanoparticles (IONPs) coated with biocompatible molecules like dimercaptosuccinic acid (DMSA), 3-aminopropyl triethoxysilane (APS) or carboxydextran (FeraSpin™ R), as well as iron oxyhydroxide nanoparticles (IOHNPs) coated with sucrose (Venofer®), or iron salts (ferric ammonium citrate -FAC), to treat and/or prevent SARS-CoV-2 infection. At non-cytotoxic doses, IONPs and IOHNPs impaired virus replication and transcription, and the production of infectious viruses in vitro, either when the cells were treated prior to or after infection, although with different efficiencies. Moreover, our data suggest that SARS-CoV-2 infection affects the expression of genes involved in cellular iron metabolism. Furthermore, the treatment of cells with IONPs and IOHNPs affects oxidative stress and iron metabolism to different extents, likely influencing virus replication and production. Interestingly, some of the nanoparticles used in this work have already been approved for their use in humans as anti-anemic treatments, such as the IOHNP Venofer®, and as contrast agents for magnetic resonance imaging in small animals like mice, such as the FeraSpin™ R IONP.
Conclusions: Therefore, our results suggest that IONPs and IOHNPs may be repurposed to be used as prophylactic or therapeutic treatments in order to combat SARS-CoV-2 infection.
Keywords: Anti-anemic; Iron metabolism; Iron oxide nanoparticles; Iron oxyhydroxide nanoparticles; MRI contrast agents; Oxidative stress; SARS-CoV-2; Viral infection; Viral replication.
© 2022. The Author(s).
Conflict of interest statement
The authors have no competing financial interests to declare.
Figures




References
MeSH terms
Substances
Grants and funding
- CSIC-COV19-012/012202020E154/Spanish National Research Council Interdisciplinary Thematic Platform (PTI) Global Health (PTI Salud Global)
- CSIC-COV19-012/012202020E154/Spanish National Research Council Interdisciplinary Thematic Platform (PTI) Global Health (PTI Salud Global)
- CSIC-COV19-012/012202020E154/Spanish National Research Council Interdisciplinary Thematic Platform (PTI) Global Health (PTI Salud Global)
- CSIC-COV19-012/012202020E154/Spanish National Research Council Interdisciplinary Thematic Platform (PTI) Global Health (PTI Salud Global)
- CSIC-COV19-012/012202020E154/Spanish National Research Council Interdisciplinary Thematic Platform (PTI) Global Health (PTI Salud Global)
- SGL2103021/European Commission-NextGenerationEU (Regulation EU2020/2094) through CSIC's Global Health Platform (PTI Salud Global)
- SGL2103021/European Commission-NextGenerationEU (Regulation EU2020/2094) through CSIC's Global Health Platform (PTI Salud Global)
- SGL2103021/European Commission-NextGenerationEU (Regulation EU2020/2094) through CSIC's Global Health Platform (PTI Salud Global)
- SGL2103021/European Commission-NextGenerationEU (Regulation EU2020/2094) through CSIC's Global Health Platform (PTI Salud Global)
- SGL2103021/European Commission-NextGenerationEU (Regulation EU2020/2094) through CSIC's Global Health Platform (PTI Salud Global)
- PDC2021-120759-100/MCIN/AEI/10.13039/501100011033 and by the "European Union NextGenerationEU/PRTR
- PDC2021-120759-100/MCIN/AEI/10.13039/501100011033 and by the "European Union NextGenerationEU/PRTR
- PDC2021-120759-100/MCIN/AEI/10.13039/501100011033 and by the "European Union NextGenerationEU/PRTR
- PDC2021-120759-100/MCIN/AEI/10.13039/501100011033 and by the "European Union NextGenerationEU/PRTR
- 2017-T1/BMD-5155/"Atracción de Talento Investigador" programme , Comunidad de Madrid
- FPU15/06170/MCIN/AEI/10.13039/501100011033 and by "ESF Investing in your future" predoctoral FPU scholar
- FPU18/04828/MCIN/AEI/10.13039/501100011033 and by "ESF Investing in your future", predoctoral FPU scholar
- JAEINT-20-01805/Spanish National Research Council JAE-INTRO 2020 Fellowship
- IJCI-2017-31447/MCIN/AEI/10.13039/501100011033, Juan de La Cierva-Incorporación Contract
- PID2020-112685RB-100/MCIN/AEI/10.13039/501100011033
LinkOut - more resources
Full Text Sources
Miscellaneous

