Host-microbiota interaction-mediated resistance to inflammatory bowel disease in pigs
- PMID: 35907917
- PMCID: PMC9338544
- DOI: 10.1186/s40168-022-01303-1
Host-microbiota interaction-mediated resistance to inflammatory bowel disease in pigs
Abstract
Background: Disease resistance phenotypes are associated with immune regulatory functions and immune tolerance and have implications for both the livestock industry and human health. Microbiota plays an essential role in regulating immunity and autoimmunity in the host organism, but the influence of host-microbiota interactions on disease resistance phenotypes remains unclear. Here, multiomics analysis was performed to identify potential regulatory mechanisms of disease resistance at both the microbiome and host levels in two pig breeds.
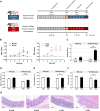
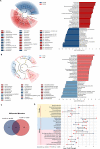
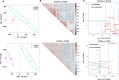
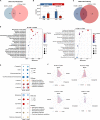
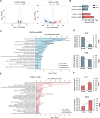
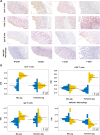
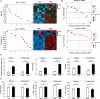
Results: Acute colitis models were established in Min pigs and Yorkshire pigs, and control and diseased individuals were compared. Compared with Yorkshire pigs under the same nutritional and management conditions, Min pigs exhibited strong disease resistance, as indicated by a low disease activity index (DAI) and a low histological activity index (HAI). Microbiota sequencing analysis showed that potentially harmful microbes Desulfovibrio, Bacteroides and Streptococcus were enriched in diseased individuals of the two breeds. Notably, potentially beneficial microbes, such as Lactobacillus, Clostridia and Eubacterium, and several genera belonging to Ruminococcaceae and Christensenellaceae were enriched in diseased Min pigs and were found to be positively associated with the microbial metabolites related to intestinal barrier function. Specifically, the concentrations of indole derivatives and short-chain fatty acids were increased in diseased Min pigs, suggesting beneficial action in protecting intestinal barrier. In addition, lower concentrations of bile acid metabolites and short-chain fatty acids were observed in diseased Yorkshire pigs, which were associated with increased potentially harmful microbes, such as Bilophila and Alistipes. Concerning enrichment of the immune response, the increase in CD4+ T cells in the lamina propria improved supervision of the host immunity response in diseased Min pigs, contributing to the maintenance of Th2-type immune superiority and immune tolerance patterns and control of excessive inflammation with the help of potentially beneficial microbes. In diseased Yorkshire pigs, more terms belonging to biological processes of immunity were enriched, including Toll-like receptors signalling, NF-κB signalling and Th1 and Th17-type immune responses, along with the increases of potentially harmful microbes and damaged intestinal barrier.
Conclusions: Cumulatively, the results for the two pig breeds highlight that host-microbiota crosstalk promotes a disease resistance phenotype in three ways: by maintaining partial PRR nonactivation, maintaining Th2-type immune superiority and immunological tolerance patterns and recovering gut barrier function to protect against colonic diseases. Video abstract.
Keywords: Disease resistance; Immunity response; Intestinal barrier function; Metabolome; Microbiota.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. - DOI - PMC - PubMed
-
- Chen LM, Collij V, Jaeger M, van den Munckhof ICL, Vila AV, Kurilshikov A, Gacesa R, Sinha T, Oosting M, Joosten LAB, et al. Gut microbial co-abundance networks show specificity in inflammatory bowel disease and obesity. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-019-13993-7. - DOI - PMC - PubMed
-
- Ijssennagger N, Belzer C, Hooiveld GJ, Dekker J, van Mil SW, Muller M, Kleerebezem M, van der Meer R. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc Natl Acad Sci U S A. 2015;112:10038–10043. doi: 10.1073/pnas.1507645112. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

