Full-length transcriptomic analysis in murine and human heart reveals diversity of PGC-1α promoters and isoforms regulated distinctly in myocardial ischemia and obesity
- PMID: 35907957
- PMCID: PMC9338484
- DOI: 10.1186/s12915-022-01360-w
Full-length transcriptomic analysis in murine and human heart reveals diversity of PGC-1α promoters and isoforms regulated distinctly in myocardial ischemia and obesity
Abstract
Background: Peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) acts as a transcriptional coactivator and regulates mitochondrial function. Various isoforms are generated by alternative splicing and differentially regulated promoters. In the heart, total PGC-1α deficiency knockout leads to dilatative cardiomyopathy, but knowledge on the complexity of cardiac isoform expression of PGC-1α remains sparse. Thus, this study aims to generate a reliable dataset on cardiac isoform expression pattern by long-read mRNA sequencing, followed by investigation of differential regulation of PGC-1α isoforms under metabolic and ischemic stress, using high-fat-high-sucrose-diet-induced obesity and a murine model of myocardial infarction.
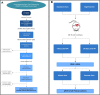
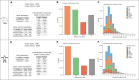
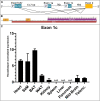
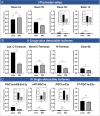
Results: Murine (C57Bl/6J) or human heart tissue (obtained during LVAD-surgery) was used for long-read mRNA sequencing, resulting in full-length transcriptomes including 58,000 mRNA isoforms with 99% sequence accuracy. Automatic bioinformatic analysis as well as manual similarity search against exonic sequences leads to identification of putative coding PGC-1α isoforms, validated by PCR and Sanger sequencing. Thereby, 12 novel transcripts generated by hitherto unknown splicing events were detected. In addition, we postulate a novel promoter with homologous and strongly conserved sequence in human heart. High-fat diet as well as ischemia/reperfusion (I/R) injury transiently reduced cardiac expression of PGC-1α isoforms, with the most pronounced effect in the infarcted area. Recovery of PGC-1α-isoform expression was even more decelerated when I/R was performed in diet-induced obese mice.
Conclusions: We deciphered for the first time a complete full-length transcriptome of the murine and human heart, identifying novel putative PGC-1α coding transcripts including a novel promoter. These transcripts are differentially regulated in I/R and obesity suggesting transcriptional regulation and alternative splicing that may modulate PGC-1α function in the injured and metabolically challenged heart.
Keywords: Diet-induced obesity; Ischemia/reperfusion; Long-read sequencing; PGC-1α.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Martínez-Redondo V, Jannig PR, Correia JC, Ferreira DM, Cervenka I, Lindvall JM, et al. Peroxisome proliferator-activated receptor γ coactivator-1 α isoforms selectively regulate multiple splicing events on target genes. J Biol Chem. 2016;291(29):15169–15184. doi: 10.1074/jbc.M115.705822. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

