The gut microbiome variability of a butterflyfish increases on severely degraded Caribbean reefs
- PMID: 35908086
- PMCID: PMC9338936
- DOI: 10.1038/s42003-022-03679-0
The gut microbiome variability of a butterflyfish increases on severely degraded Caribbean reefs
Abstract
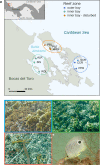
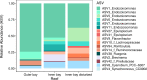
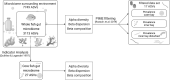
Environmental degradation has the potential to alter key mutualisms that underlie the structure and function of ecological communities. How microbial communities associated with fishes vary across populations and in relation to habitat characteristics remains largely unknown despite their fundamental roles in host nutrition and immunity. We find significant differences in the gut microbiome composition of a facultative coral-feeding butterflyfish (Chaetodon capistratus) across Caribbean reefs that differ markedly in live coral cover (∼0-30%). Fish gut microbiomes were significantly more variable at degraded reefs, a pattern driven by changes in the relative abundance of the most common taxa potentially associated with stress. We also demonstrate that fish gut microbiomes on severely degraded reefs have a lower abundance of Endozoicomonas and a higher diversity of anaerobic fermentative bacteria, which may suggest a less coral dominated diet. The observed shifts in fish gut bacterial communities across the habitat gradient extend to a small set of potentially beneficial host associated bacteria (i.e., the core microbiome) suggesting essential fish-microbiome interactions may be vulnerable to severe coral degradation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Idjadi J, Edmunds P. Scleractinian corals as facilitators for other invertebrates on a Caribbean reef. Mar. Ecol. Prog. Ser. 2006;319:117–127. doi: 10.3354/meps319117. - DOI
-
- Norström A, Nyström M, Lokrantz J, Folke C. Alternative states on coral reefs: beyond coral–macroalgal phase shifts. Mar. Ecol. Prog. Ser. 2009;376:295–306. doi: 10.3354/meps07815. - DOI
-
- Wilson SK, Graham NAJ, Pratchett MS, Jones GP, Polunin NVC. Multiple disturbances and the global degradation of coral reefs: are reef fishes at risk or resilient? Glob. Chang. Biol. 2006;12:2220–2234. doi: 10.1111/j.1365-2486.2006.01252.x. - DOI

