Theaflavin 3-gallate inhibits the main protease (Mpro) of SARS-CoV-2 and reduces its count in vitro
- PMID: 35908093
- PMCID: PMC9338964
- DOI: 10.1038/s41598-022-17558-5
Theaflavin 3-gallate inhibits the main protease (Mpro) of SARS-CoV-2 and reduces its count in vitro
Abstract
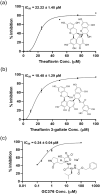
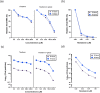
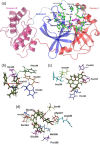
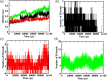
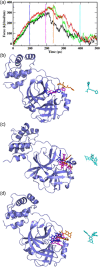
The main protease (Mpro) of SARS-CoV-2 has been recognized as an attractive drug target because of its central role in viral replication. Our previous preliminary molecular docking studies showed that theaflavin 3-gallate (a natural bioactive molecule derived from theaflavin and found in high abundance in black tea) exhibited better docking scores than repurposed drugs (Atazanavir, Darunavir, Lopinavir). In this study, conventional and steered MD-simulations analyses revealed stronger interactions of theaflavin 3-gallate with the active site residues of Mpro than theaflavin and a standard molecule GC373 (a known inhibitor of Mpro and novel broad-spectrum anti-viral agent). Theaflavin 3-gallate inhibited Mpro protein of SARS-CoV-2 with an IC50 value of 18.48 ± 1.29 μM. Treatment of SARS-CoV-2 (Indian/a3i clade/2020 isolate) with 200 μM of theaflavin 3-gallate in vitro using Vero cells and quantifying viral transcripts demonstrated reduction of viral count by 75% (viral particles reduced from Log106.7 to Log106.1). Overall, our findings suggest that theaflavin 3-gallate effectively targets the Mpro thus limiting the replication of the SARS-CoV-2 virus in vitro.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Black tea bioactives as inhibitors of multiple targets of SARS-CoV-2 (3CLpro, PLpro and RdRp): a virtual screening and molecular dynamic simulation study.J Biomol Struct Dyn. 2022 Sep;40(15):7143-7166. doi: 10.1080/07391102.2021.1897679. Epub 2021 Mar 10. J Biomol Struct Dyn. 2022. PMID: 33715595
-
Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors.J Biomol Struct Dyn. 2021 Jul;39(10):3449-3458. doi: 10.1080/07391102.2020.1766572. Epub 2020 May 20. J Biomol Struct Dyn. 2021. PMID: 32397940 Free PMC article.
-
Theaflavin-3'-O-gallate a Black-tea Constituent Blocked SARS CoV-2 RNA dependant RNA Polymerase Active-site with Better Docking Results than Remdesivir.Drug Res (Stuttg). 2021 Oct;71(8):462-472. doi: 10.1055/a-1467-5828. Epub 2021 Sep 13. Drug Res (Stuttg). 2021. PMID: 34517419
-
Repurposing an Antiviral Drug against SARS-CoV-2 Main Protease.Angew Chem Int Ed Engl. 2021 Oct 25;60(44):23492-23494. doi: 10.1002/anie.202107481. Epub 2021 Sep 21. Angew Chem Int Ed Engl. 2021. PMID: 34545983 Free PMC article. Review.
-
An Updated Review on SARS-CoV-2 Main Proteinase (MPro): Protein Structure and Small-Molecule Inhibitors.Curr Top Med Chem. 2021;21(6):442-460. doi: 10.2174/1568026620666201207095117. Curr Top Med Chem. 2021. PMID: 33292134 Review.
Cited by
-
Potential anti-hyperglycemic activity of black tea theaflavins through inhibiting α-amylase.Food Chem X. 2024 Mar 16;22:101296. doi: 10.1016/j.fochx.2024.101296. eCollection 2024 Jun 30. Food Chem X. 2024. PMID: 38550892 Free PMC article.
-
Machine learning combines atomistic simulations to predict SARS-CoV-2 Mpro inhibitors from natural compounds.Mol Divers. 2024 Apr;28(2):553-561. doi: 10.1007/s11030-023-10601-1. Epub 2023 Feb 24. Mol Divers. 2024. PMID: 36823394 Free PMC article.
-
Ginkgolic acids inhibit SARS-CoV-2 and its variants by blocking the spike protein/ACE2 interplay.Int J Biol Macromol. 2023 Jan 31;226:780-792. doi: 10.1016/j.ijbiomac.2022.12.057. Epub 2022 Dec 12. Int J Biol Macromol. 2023. PMID: 36521705 Free PMC article.
-
Targeting the SARS-CoV-2 HR1 with Small Molecules as Inhibitors of the Fusion Process.Int J Mol Sci. 2022 Sep 3;23(17):10067. doi: 10.3390/ijms231710067. Int J Mol Sci. 2022. PMID: 36077465 Free PMC article.
-
Antiviral efficacy of honey bee antimicrobial peptides against SARS-CoV-2.Mol Divers. 2025 Aug 16. doi: 10.1007/s11030-025-11325-0. Online ahead of print. Mol Divers. 2025. PMID: 40819116
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

