N/C Interactions Are Dispensable for Normal In Vivo Functioning of the Androgen Receptor in Male Mice
- PMID: 35908178
- PMCID: PMC9756762
- DOI: 10.1210/endocr/bqac104
N/C Interactions Are Dispensable for Normal In Vivo Functioning of the Androgen Receptor in Male Mice
Abstract
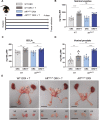
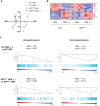
The androgen receptor (AR) plays a central role in the development and maintenance of the male phenotype. The binding of androgens to the receptor induces interactions between the carboxyterminal ligand-binding domain and the highly conserved 23FQNLF27 motif in the aminoterminal domain. The role of these so-called N/C interactions in AR functioning is debated. In vitro assays show that mutating the AR in the 23FQNLF27 motif (called ARNoC) attenuates the AR transactivation of reporter genes, has no effect on ligand binding, but does affect protein-protein interactions with several AR coregulators. To test the in vivo relevance of the N/C interaction, we analyzed the consequences of the genomic introduction of the ARNoC mutation in mice. Surprisingly, the ARNoC/Y mice show a normal male development, with unaffected male anogenital distance and normal accessory sex glands, male circulating androgen levels, body composition, and fertility. The responsiveness of androgen target genes in kidney, prostate, and testes was also unaffected. We thus conclude that the N/C interactions in the AR are not essential for the development of a male phenotype under normal physiological conditions.
Keywords: N/C interaction; androgen receptor; male development; mouse model; prostate.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Endocrine Society. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
The authors declare no competing interests.
The authors have nothing to disclose.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

