Glioblastoma-educated mesenchymal stem-like cells promote glioblastoma infiltration via extracellular matrix remodelling in the tumour microenvironment
- PMID: 35908277
- PMCID: PMC9339241
- DOI: 10.1002/ctm2.997
Glioblastoma-educated mesenchymal stem-like cells promote glioblastoma infiltration via extracellular matrix remodelling in the tumour microenvironment
Abstract
Background: The biological function of mesenchymal stem-like cells (MSLCs), a type of stromal cells, in the regulation of the tumour microenvironment is unclear. Here, we investigated the molecular mechanisms underlying extracellular matrix (ECM) remodelling and crosstalk between MSLCs and glioblastomas (GBMs) in tumour progression.
Methods: In vitro and in vivo co-culture systems were used to analyze ECM remodelling and GBM infiltration. In addition, clinical databases, samples from patients with GBM and a xenografted mouse model of GBM were used.
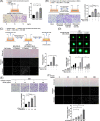
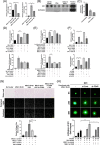
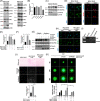
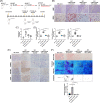
Results: Previous studies have shown that the survival of patients with GBM from whom MSLCs could be isolated is substantially shorter than that of patients from whom MSLCs could not be isolated. Therefore, we determined the correlation between changes in ECM-related gene expression in MSLC-isolatable patients with that in MSLC non-isolatable patients using gene set enrichment analysis (GSEA). We found that lysyl oxidase (LOX) and COL1A1 expressions increased in MSLCs via GBM-derived clusters of differentiation 40 ligand (CD40L). Mechanistically, MSLCs are reprogrammed by the CD40L/CD40/NFκB2 signalling axis to build a tumour infiltrative microenvironment involving collagen crosslinking. Importantly, blocking of CD40L by a neutralizing antibody-suppressed LOX expression and ECM remodelling, decreasing GBM infiltration in mouse xenograft models. Clinically, high expression of CD40L, clusters of differentiation 40 (CD40) and LOX correlated with poor survival in patients with glioma. This indicated that GBM-educated MSLCs promote GBM infiltration via ECM remodelling in the tumour microenvironment.
Conclusion: Our findings provide mechanistic insights into the pro-infiltrative tumour microenvironment produced by GBM-educated MSLCs and highlight a potential therapeutic target that can be used for suppressing GBM infiltration.
Keywords: cluster of differentiation 40 ligand; extracellular matrix remodelling; glioblastoma; lysyl oxidase; mesenchymal stem-like cells.
© 2022 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare that there is no potential conflict of interest.
Figures


Similar articles
-
Crosstalk between GBM cells and mesenchymal stemlike cells promotes the invasiveness of GBM through the C5a/p38/ZEB1 axis.Neuro Oncol. 2020 Oct 14;22(10):1452-1462. doi: 10.1093/neuonc/noaa064. Neuro Oncol. 2020. PMID: 32179921 Free PMC article.
-
Increased in vivo angiogenic effect of glioma stromal mesenchymal stem-like cells on glioma cancer stem cells from patients with glioblastoma.Int J Oncol. 2013 May;42(5):1754-62. doi: 10.3892/ijo.2013.1856. Epub 2013 Mar 12. Int J Oncol. 2013. PMID: 23483121
-
The mode and dynamics of glioblastoma cell invasion into a decellularized tissue-derived extracellular matrix-based three-dimensional tumor model.Sci Rep. 2018 Mar 15;8(1):4608. doi: 10.1038/s41598-018-22681-3. Sci Rep. 2018. PMID: 29545552 Free PMC article.
-
Glioblastoma Microenvironment and Invasiveness: New Insights and Therapeutic Targets.Int J Mol Sci. 2023 Apr 11;24(8):7047. doi: 10.3390/ijms24087047. Int J Mol Sci. 2023. PMID: 37108208 Free PMC article. Review.
-
Glioblastoma mechanobiology at multiple length scales.Biomater Adv. 2024 Jun;160:213860. doi: 10.1016/j.bioadv.2024.213860. Epub 2024 Apr 15. Biomater Adv. 2024. PMID: 38640876 Review.
Cited by
-
Identification of lysyl oxidase as an adipocyte-secreted mediator that promotes a partial mesenchymal-to-epithelial transition in MDA-MB-231 cells.Explor Target Antitumor Ther. 2024;5(1):1-19. doi: 10.37349/etat.2024.00201. Epub 2024 Jan 16. Explor Target Antitumor Ther. 2024. PMID: 38468823 Free PMC article.
-
Speeding up Glioblastoma Cancer Research: Highlighting the Zebrafish Xenograft Model.Int J Mol Sci. 2024 May 15;25(10):5394. doi: 10.3390/ijms25105394. Int J Mol Sci. 2024. PMID: 38791432 Free PMC article. Review.
-
Identification of LOX as a candidate prognostic biomarker in Glioblastoma multiforme.Transl Oncol. 2023 Oct;36:101739. doi: 10.1016/j.tranon.2023.101739. Epub 2023 Aug 4. Transl Oncol. 2023. PMID: 37544033 Free PMC article.
-
HAS2 facilitates glioma cell malignancy and suppresses ferroptosis in an FZD7-dependent manner.Cancer Sci. 2024 Aug;115(8):2602-2616. doi: 10.1111/cas.16232. Epub 2024 May 30. Cancer Sci. 2024. PMID: 38816349 Free PMC article.
-
Key Cell-in-Cell Related Genes are Identified by Bioinformatics and Experiments in Glioblastoma.Cancer Manag Res. 2024 Sep 5;16:1109-1130. doi: 10.2147/CMAR.S475513. eCollection 2024. Cancer Manag Res. 2024. PMID: 39253064 Free PMC article.
References
-
- Stummer W, Pichlmeier U, Meinel T, Wiestler OD, Zanella F, Reulen HJ. Fluorescence‐guided surgery with 5‐aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7(5):392‐401. - PubMed
-
- Caverzasi E, SL Hervey‐Jumper, Jordan KM, et al. Identifying preoperative language tracts and predicting postoperative functional recovery using HARDI q‐ball fiber tractography in patients with gliomas. J Neurosurg. 2016;125(1):33‐45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
