A redox-sensitive iron-sulfur cluster in murine FAM72A controls its ability to degrade the nuclear form of uracil-DNA glycosylase
- PMID: 35908367
- PMCID: PMC10996437
- DOI: 10.1016/j.dnarep.2022.103381
A redox-sensitive iron-sulfur cluster in murine FAM72A controls its ability to degrade the nuclear form of uracil-DNA glycosylase
Abstract
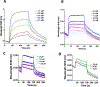
Murine FAM72A, mFAM72A, binds the nuclear form of uracil-DNA glycosylase, mUNG2, inhibits its activity and causes its degradation. In immunoprecipitation assays the human paralog, hFAM72A, binds hUNG2 and is a potential anti-cancer drug target because of its high expression in many cancers. Using purified mFAM72A, and mUNG2 proteins we show that mFAM72A binds mUNG2, and the N-terminal 25 amino acids of mUNG2 bind mFAM72A at a nanomolar dissociation constant. We also show that mFAM72A is present throughout the cells, and mUNG2 helps localize it to nuclei. Based on in silico models of mFAM72A-mUNG2 interactions, we constructed several mutants of mFAM72A and found that while they have reduced ability to deplete mUNG2, the mutations also destabilized the former protein. We confirmed that Withaferin A, a predicted lead molecule for the design of FAM72A inhibitors, binds mFAM72A with micromolar affinity but has little affinity to mUNG2. We identified two potential metal-binding sites in mFAM72A and show that one of the sites contains an Fe-S cluster. This redox-sensitive cluster is involved in the mFAM72A-mUNG2 interaction and modulates mFAM72A activity. Hydrogen peroxide treatment of cells increases mUNG2 depletion in a FAM72A-dependent fashion suggesting that mFAM72A activity is redox-sensitive.
Keywords: Class-switch recombination; Iron-sulfur cluster; Oxidative stress; Somatic hypermutation; Uracil-DNAglycosylase.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that there are no conflicts of interest.
Figures




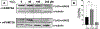
References
-
- Heese K, The protein p17 signaling pathways in cancer, Tumour Biol, 34 (2013) 4081–4087. - PubMed
-
- Wang LT, Lin CS, Chai CY, Liu KY, Chen JY, Hsu SH, Functional interaction of Ugene and EBV infection mediates tumorigenic effects, Oncogene, 30 (2011) 2921–2932. - PubMed
-
- Rajan P, Stockley J, Sudbery IM, Fleming JT, Hedley A, Kalna G, Sims D, Ponting CP, Heger A, Robson CN, McMenemin RM, Pedley ID, Leung HY, Identification of a candidate prognostic gene signature by transcriptome analysis of matched pre- and post-treatment prostatic biopsies from patients with advanced prostate cancer, BMC Cancer, 14 (2014) 977. - PMC - PubMed
-
- Rahane CS, Kutzner A, Heese K, A cancer tissue-specific FAM72 expression profile defines a novel glioblastoma multiform (GBM) gene-mutation signature, J Neurooncol, 141 (2019) 57–70. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

