Loss of mitochondrial enzyme GPT2 causes early neurodegeneration in locus coeruleus
- PMID: 35908744
- PMCID: PMC9669404
- DOI: 10.1016/j.nbd.2022.105831
Loss of mitochondrial enzyme GPT2 causes early neurodegeneration in locus coeruleus
Abstract
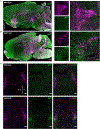
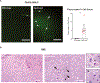
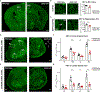
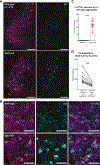
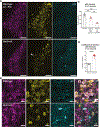
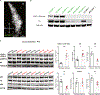
Locus coeruleus (LC) is among the first brain areas to degenerate in Alzheimer's disease and Parkinson's disease; however, the underlying causes for the vulnerability of LC neurons are not well defined. Here we report a novel mechanism of degeneration of LC neurons caused by loss of the mitochondrial enzyme glutamate pyruvate transaminase 2 (GPT2). GPT2 Deficiency is a newly-recognized childhood neurometabolic disorder. The GPT2 enzyme regulates cell growth through replenishment of tricarboxylic acid (TCA) cycle intermediates and modulation of amino acid metabolism. In Gpt2-null mice, we observe an early loss of tyrosine hydroxylase (TH)-positive neurons in LC and reduced soma size at postnatal day 18. Gpt2-null LC shows selective positive Fluoro-Jade C staining. Neuron loss is accompanied by selective, prominent microgliosis and astrogliosis in LC. We observe reduced noradrenergic projections to and norepinephrine levels in hippocampus and spinal cord. Whole cell recordings in Gpt2-null LC slices show reduced soma size and abnormal action potentials with altered firing kinetics. Strikingly, we observe early decreases in phosphorylated S6 in Gpt2-null LC, preceding prominent p62 aggregation, increased LC3B-II to LC3B-I ratio, and neuronal loss. These data are consistent with a possible mechanism involving deficiency in protein synthesis and cell growth, associated subsequently with abnormal autophagy and neurodegeneration. As compared to the few genetic animal models with LC degeneration, loss of LC neurons in Gpt2-null mice is developmentally the earliest. Early neuron loss in LC in a model of human neurometabolic disease provides important clues regarding the metabolic vulnerability of LC and may lead to new therapeutic targets.
Keywords: Autophagy; GPT2; Locus coeruleus; Neurodegeneration; Neurogenetics; Neurometabolism; Proteostasis; Selective vulnerability.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no competing interests.
Figures

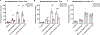
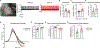
References
-
- Amaral D, Sinnamon H, 1977. The locus coeruleus: neurobiology of a central noradrenergic nucleus. Prog. Neurobiol 9, 147–196. - PubMed
-
- Aston-Jones G, Rajkowski J, Cohen J, 1999. Role of locus coeruleus in attention and behavioral flexibility. Biol. Psychiatry 46, 1309–1320. - PubMed
-
- Bartlett BJ, Isakson P, Lewerenz J, Sanchez H, Kotzebue RW, Cumming RC, Harris GL, Nezis IP, Schubert DR, Simonsen A, et al. , 2011. p62, ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 7, 572–583. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

