Hsa_circ_0129047 regulates the miR-375/ACVRL1 axis to attenuate the progression of lung adenocarcinoma
- PMID: 35908770
- PMCID: PMC9459267
- DOI: 10.1002/jcla.24591
Hsa_circ_0129047 regulates the miR-375/ACVRL1 axis to attenuate the progression of lung adenocarcinoma
Abstract
Background: Circular RNAs (circRNAs) are attractive candidates to be used as biomarkers of human cancers, including lung adenocarcinoma (LUAD). Our study aimed to investigate the functions and regulatory mechanisms of hsa_circ_0129047 in the tumorigenesis of LUAD.
Methods: Reverse transcription-quantitative polymerase chain reaction was performed to determine the circRNA, microRNA (miRNA), and mRNA expression levels in LUAD cell lines and tissues. Tumor xenografts were established in nude mice to evaluate whether hsa_circ_0129047 affected LUAD tumor development in vivo. Cell counting kit-8 and transwell assays were performed to assess the mechanisms by which hsa_circ_0129047 influenced the viability and migration of LUAD cells, respectively. Apoptosis was evaluated via determination of the levels of the apoptotic markers, B-cell lymphoma-2, and Bcl-2-associated X, via Western blotting. Dual-luciferase reporter assay, RNA immunoprecipitation assay, and Pearson's correlation analysis were performed to determine the relationships among miR-375 and hsa_circ_0129047 and activin A receptor-like type 1 (ACVRL1).
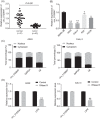
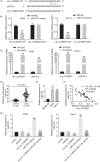
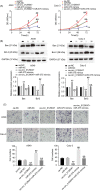
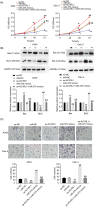
Results: Downregulation of hsa_circ_0129047 levels was observed in LUAD cell lines and tissues. Meanwhile, the upregulation of hsa_circ_0129047 levels repressed the proliferative, migratory, and survival capacities of LUAD cells in vitro. Hsa_circ_0129047 exerted antitumor effects during in vivo tumor development. Finally, we demonstrated that hsa_circ_0129047 sponged miR-375. This interaction facilitated the expression of the downstream target of miR-375, ACVRL1, whose upregulation inhibited the development and malignancy of LUAD.
Conclusion: These findings demonstrate that hsa_circ_0129047 functions as a tumor inhibitor in LUAD by modulating the miR-375/ACVRL1 axis. Hence, hsa_circ_0129047 may be a promising biomarker and gene target for LUAD treatment.
Keywords: ACVRL1; hsa_circ_0129047; lung adenocarcinoma; miR-375.
© 2022 The Authors. Journal of Clinical Laboratory Analysis published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675‐691. - PubMed
-
- Chen B, Huang S. Circular RNA: an emerging non‐coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41‐50. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

