Predicting of molecules mediating an interaction between bovine embryos and uterine epithelial cells
- PMID: 35908976
- PMCID: PMC9558814
- DOI: 10.1262/jrd.2022-046
Predicting of molecules mediating an interaction between bovine embryos and uterine epithelial cells
Abstract
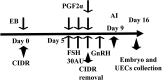
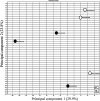
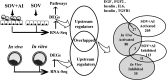
Embryo-maternal reproductive tract interactions are pivotal for successful pregnancy. The present study predicted the molecules modulating embryo-uterine communication by comparing two sets of differentially expressed genes (DEGs): DEGs in uterine epithelial cells (UECs) collected from the uterus with and without blastocysts and DEGs between blastocysts developed in vivo and in vitro. Cows were subjected to super ovulation (SOV), followed by insemination or non-insemination at estrus (SOV + AI and SOV cows). Seven days after estrus, the uterus was flushed to collect UECs, and the presence of blastocysts in the uterus was confirmed. UECs were subjected to RNA-Sequencing (RNA-Seq) to identify DEGs. Publicly available RNA-Seq data of in vivo and in vitro developed bovine blastocysts were used to determine DEGs. Then, using ingenuity pathway analysis, activated- and inhibited-upstream regulators (USRs) for UECs in blastocysts were compared with those for blastocysts developed in vivo. RNA-Seq of UECs revealed that the DEGs were associated with immune response and cell adhesion pathways. The activated and inhibited USRs of UECs derived from SOV+ AI cows overlapped with the activated and inhibited USRs of blastocysts developed in vivo. Overlapping activated USRs include leukemia inhibitory factor, interleukin 6, fibroblast growth factor-2, transforming growth factor beta-1, and epidermal growth factor. In conclusion, the present study predicted the molecules that potentially mediate communication between the developing embryo and the uterus in vivo and prepare the uterus for pregnancy.
Keywords: Blastocyst; Gene expression; Ingenuity pathway analysis; Upstream regulators; Uterine epithelial cells.
Conflict of interest statement
We declare no conflicts of interest in this study.
Figures
Similar articles
-
Expression profile of adhesion molecules in blastocyst vis-a-vis uterine epithelial cells.Theriogenology. 2021 Aug;170:36-45. doi: 10.1016/j.theriogenology.2021.04.018. Epub 2021 May 5. Theriogenology. 2021. PMID: 33984621
-
Bovine embryo culture in the presence or absence of serum: implications for blastocyst development, cryotolerance, and messenger RNA expression.Biol Reprod. 2003 Jan;68(1):236-43. doi: 10.1095/biolreprod.102.007799. Biol Reprod. 2003. PMID: 12493719
-
Effects of intrauterine extracellular vesicle microRNAs on embryonic gene expression in low-fertility cows.FASEB J. 2024 Oct 31;38(20):e70116. doi: 10.1096/fj.202401728R. FASEB J. 2024. PMID: 39425543 Free PMC article.
-
Oocyte maturation under lipotoxic conditions induces carryover transcriptomic and functional alterations during post-hatching development of good-quality blastocysts: novel insights from a bovine embryo-transfer model.Hum Reprod. 2020 Feb 29;35(2):293-307. doi: 10.1093/humrep/dez248. Hum Reprod. 2020. PMID: 32112081
-
Progesterone modulates adhesion molecules in uterine epithelial cells and in vitro embryo production in buffalo.Reprod Domest Anim. 2020 Jul;55(7):833-843. doi: 10.1111/rda.13691. Epub 2020 May 18. Reprod Domest Anim. 2020. PMID: 32335951
Cited by
-
Simple and Efficient Chemically Defined In Vitro Maturation and Embryo Culture System for Bovine Embryos.Animals (Basel). 2022 Nov 7;12(21):3057. doi: 10.3390/ani12213057. Animals (Basel). 2022. PMID: 36359181 Free PMC article.
-
Beneficial Effect of Polysaccharide Gel Made of Xanthan Gum and Locust Bean Gum on Bovine Oocytes.Int J Mol Sci. 2023 Feb 9;24(4):3508. doi: 10.3390/ijms24043508. Int J Mol Sci. 2023. PMID: 36834915 Free PMC article.
-
Genotyping of cows by LHCGR, FSHR loci, and determination of the level of ovulation depending on the expression of the studied genes.Open Vet J. 2023 Mar;13(3):352-357. doi: 10.5455/OVJ.2023.v13.i3.12. Epub 2023 Mar 18. Open Vet J. 2023. PMID: 37026064 Free PMC article.

