Decoding the roles of extremophilic microbes in the anaerobic environments: Past, Present, and Future
- PMID: 35909618
- PMCID: PMC9325894
- DOI: 10.1016/j.crmicr.2022.100146
Decoding the roles of extremophilic microbes in the anaerobic environments: Past, Present, and Future
Abstract
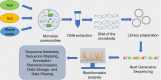
The genome of an organism is directly or indirectly correlated with its environment. Consequently, different microbes have evolved to survive and sustain themselves in a variety of environments, including unusual anaerobic environments. It is believed that their genetic material could have played an important role in the early evolution of their existence in the past. Presently, out of the uncountable number of microbes found in different ecosystems we have been able to discover only one percent of the total communities. A large majority of the microbial populations exists in the most unusual and extreme environments. For instance, many anaerobic bacteria are found in the gastrointestinal tract of humans, soil, and hydrothermal vents. The recent advancements in Metagenomics and Next Generation Sequencing technologies have improved the understanding of their roles in these environments. Presently, anaerobic bacteria are used in various industries associated with biofuels, fermentation, production of enzymes, vaccines, vitamins, and dairy products. This broad applicability brings focus to the significant contribution of their genomes in these functions. Although the anaerobic microbes have become an irreplaceable component of our lives, a major and important section of such anaerobic microbes still remain unexplored. Therefore, it can be said that unlocking the role of the microbial genomes of the anaerobes can be a noteworthy discovery not just for mankind but for the entire biosystem as well.
Keywords: Anaerobe; Anaerobic; Extreme environments; Metagenomics; Microbes.
© 2022 The Author(s). Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Figures
Similar articles
-
Dynamics and genetic diversification of Escherichia coli during experimental adaptation to an anaerobic environment.PeerJ. 2017 May 3;5:e3244. doi: 10.7717/peerj.3244. eCollection 2017. PeerJ. 2017. PMID: 28480139 Free PMC article.
-
Potential Interactions between Clade SUP05 Sulfur-Oxidizing Bacteria and Phages in Hydrothermal Vent Sponges.Appl Environ Microbiol. 2019 Oct 30;85(22):e00992-19. doi: 10.1128/AEM.00992-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31492669 Free PMC article.
-
Genomic Insights into the Ecological Role and Evolution of a Novel Thermoplasmata Order, "Candidatus Sysuiplasmatales".Appl Environ Microbiol. 2021 Oct 28;87(22):e0106521. doi: 10.1128/AEM.01065-21. Epub 2021 Sep 15. Appl Environ Microbiol. 2021. PMID: 34524897 Free PMC article.
-
Anaerobes as Sources of Bioactive Compounds and Health Promoting Tools.Adv Biochem Eng Biotechnol. 2016;156:433-464. doi: 10.1007/10_2016_6. Adv Biochem Eng Biotechnol. 2016. PMID: 27432247 Review.
-
Metagenomic analyses: past and future trends.Appl Environ Microbiol. 2011 Feb;77(4):1153-61. doi: 10.1128/AEM.02345-10. Epub 2010 Dec 17. Appl Environ Microbiol. 2011. PMID: 21169428 Free PMC article. Review.
Cited by
-
Response of alcohol fermentation strains, mixed fermentation and extremozymes interactions on wine flavor.Front Microbiol. 2025 Jan 29;16:1532539. doi: 10.3389/fmicb.2025.1532539. eCollection 2025. Front Microbiol. 2025. PMID: 39944646 Free PMC article. Review.
References
-
- Ahmad, A., Naqvi, S.A., Muhammad, Jaskani, J., Waseem, Muhammad, Ali, E., Khan, I.A., Manzoor, F., Siddeeg, A., Aadil, R.M., 2021. Efficient utilization of date palm waste for the bioethanol production through Saccharomyces cerevisiae strain. https://doi.org/10.1002/fsn3.2175. - PMC - PubMed
-
- Alalawy A.I., Guo Z., Almutairi F.M., El Rabey H.A., Al-Duais Mohammed A., Mohammed Ghena M., Almasoudi F.M., Alotaibi M.A., Salama E.-.S., Abomohra Abd, El-Fatah Sakran M.I. Explication of structural variations in the bacterial and archaeal community of anaerobic digestion sludges: an insight through metagenomics. J. Environ. Chem. Eng. 2021;9 doi: 10.1016/j.jece.2021.105910. - DOI
-
- Alba-Lois L., Segal-Kischinevzky C. Yeast fermentation and the making of beer and wine. Nat. Educ. 2010;3:1–4.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous