Phenotype to genotype in Neurospora crassa: Association of the scumbo phenotype with mutations in the gene encoding ceramide C9-methyltransferase
- PMID: 35909622
- PMCID: PMC9325734
- DOI: 10.1016/j.crmicr.2022.100117
Phenotype to genotype in Neurospora crassa: Association of the scumbo phenotype with mutations in the gene encoding ceramide C9-methyltransferase
Abstract
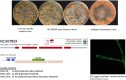
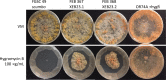
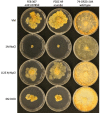
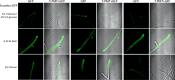
Using a legacy of genetic mutants of Neurospora crassa, paired with resequencing efforts through JGI, we have identified the gene responsible for the 'scumbo' mutant. This early morphological mutant was described as "Irregular flat, spreading growth with knobby protrusions and abnormal conidiation, but no free conidia. Mycelium usually appears yellowish rather than orange. Female fertile." (Perkins, Radford et al. 2000). Our further investigation has found new insights into the identity and associated functions of scumbo as a ceramide C9 methyltransferase, previously annotated as "similar to cyclopropane-fatty-acyl-phospholipidsynthase", encoded by the gene NCU07859. This enzyme performs a fungal-specific methyl modification of glycosyl-ceramides and has implications for membrane homeostasis and hyphal polarity in filamentous fungi.
Keywords: Ceramide C9-methyltransferase; Fast-forward genetics; Fungal genetics; Fungal genomics; Fungi; Morphology; Neurospora; Scumbo.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Abramsky T., Tatum E. Differential inhibition of branching enzyme in a morphological mutant and in wild type Neurospora - Influence of carbon source in the growth medium. Biochimica Et Biophysica Acta. 1976;421(1):106–114. - PubMed
-
- Akhberdi O., Zhang Q., Wang H., Li Y., Chen L., Wang D., Yu X., Wei D., Zhu X. Roles of phospholipid methyltransferases in pycnidia development, stress tolerance and secondary metabolism in the taxol-producing fungus Pestalotiopsis microspore. Microbiological Research. 2018;210:33–42. - PubMed
-
- Baker S.E. Selection to sequence: opportunities in fungal genomics. Environmental Microbiology. 2009;11(12):2955–2958. - PubMed
-
- Baker S.E., Bredeweg E.L. Comparative genomics, resequencing and fast forward genetics in Aspergillus and Penicillium. Aspergillus and Penicillium in the Post-genomic Era. 2016:17–26.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

