Glycogen synthase kinases in model and crop plants - From negative regulators of brassinosteroid signaling to multifaceted hubs of various signaling pathways and modulators of plant reproduction and yield
- PMID: 35909730
- PMCID: PMC9335153
- DOI: 10.3389/fpls.2022.939487
Glycogen synthase kinases in model and crop plants - From negative regulators of brassinosteroid signaling to multifaceted hubs of various signaling pathways and modulators of plant reproduction and yield
Abstract
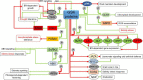
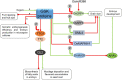
Glycogen synthase kinases, also known as SHAGGY-like Kinases (GSKs/SKs), are highly conserved serine/threonine protein kinases present both in animals and plants. Plant genomes contain multiple homologs of the GSK3 genes which participate in various biological processes. Plant GSKs/SKs, and their best known representative in Arabidopsis thaliana - Brassinosteroid Insentisive2 (BIN2/SK21) in particular, were first identified as components of the brassinosteroid (BR) signaling pathway. As phytohormones, BRs regulate a wide range of physiological processes in plants - from germination, cell division, elongation and differentiation to leaf senescence, and response to environmental stresses. The GSKs/SKs proteins belong to a group of several highly conserved components of the BR signaling which evolved early during evolution of this molecular relay. However, recent reports indicated that the GSKs/SKs proteins are also implicated in signaling pathways of other phytohormones and stress-response processes. As a consequence, the GSKs/SKs proteins became hubs of various signaling pathways and modulators of plant development and reproduction. Thus, it is very important to understand molecular mechanisms regulating activity of the GSKs/SKs proteins, but also to get insights into role of the GSKs/SKs proteins in modulation of stability and activity of various substrate proteins which participate in the numerous signaling pathways. Although elucidation of these aspects is still in progress, this review presents a comprehensive and detailed description of these processes and their implications for regulation of development, stress response, and reproduction of model and crop species. The GSKs/SKs proteins and their activity are modulated through phosphorylation and de-phosphorylation reactions which are regulated by various proteins. Importantly, both phosphorylations and de-phosphorylations may have positive and negative effects on the activity of the GSKs/SKs proteins. Additionally, the activity of the GSKs/SKs proteins is positively regulated by reactive oxygen species, whereas it is negatively regulated through ubiquitylation, deacetylation, and nitric oxide-mediated nitrosylation. On the other hand, the GSKs/SKs proteins interact with proteins representing various signaling pathways, and on the basis of the complicated network of interactions the GSKs/SKs proteins differentially regulate various physiological, developmental, stress response, and yield-related processes.
Keywords: brassinosteroids; crosstalk; glycogen synthase kinases; plant reproduction; plant yield; signaling; stress response.
Copyright © 2022 Zolkiewicz and Gruszka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ackerman-Lavert M., Savaldi-Goldstein S. (2020). Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 53 90–97. - PubMed
-
- Amorim-Silva V., García-Moreno Á., Castillo A. G., Lakhssassi N., Esteban Del Valle A., Pérez-Sancho J., et al. (2019). TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in Arabidopsis. Plant Cell 31 1807–1828. 10.1105/tpc.19.00150 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

