Genome-Wide Analysis of Anthocyanin Biosynthesis Regulatory WD40 Gene FcTTG1 and Related Family in Ficus carica L
- PMID: 35909733
- PMCID: PMC9334019
- DOI: 10.3389/fpls.2022.948084
Genome-Wide Analysis of Anthocyanin Biosynthesis Regulatory WD40 Gene FcTTG1 and Related Family in Ficus carica L
Abstract
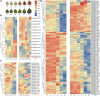
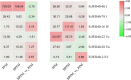
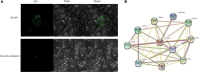
WD40 proteins serve as crucial regulators in a broad spectrum of plant developmental and physiological processes, including anthocyanin biosynthesis. However, in fig (Ficus carica L.), neither the WD40 family nor any member involved in anthocyanin biosynthesis has been elucidated. In the present study, 204 WD40 genes were identified from the fig genome and phylogenetically classified into 5 clusters and 12 subfamilies. Bioinformatics analysis prediction localized 109, 69, and 26 FcWD40 proteins to the cytoplasm, nucleus and other cellular compartments, respectively. RNA-seq data mining revealed 127 FcWD40s expressed at FPKM > 10 in fig fruit. Most of these genes demonstrated higher expression in the early stages of fruit development. FcWD40-97 was recruited according to three criteria: high expression in fig fruit, predicted nuclear localization, and closest clustering with TTG1s identified in other plants. FcWD40-97, encoding 339 amino acids including 5 WD-repeat motifs, showed 88.01 and 87.94% amino acid sequence similarity to apple and peach TTG1, respectively. The gene is located on fig chromosome 4, and is composed of 1 intron and 2 exons. Promoter analysis revealed multiple light-responsive elements, one salicylic acid-responsive element, three methyl jasmonate-responsive elements, and one MYB-binding site involved in flavonoid biosynthesis gene regulation. FcWD40-97 was in the FPKM > 100 expression level group in fig fruit, and higher expression was consistently found in the peel compared to the flesh at the same development stages. Expression level did not change significantly under light deprivation, whereas in leaves and roots, its expression was relatively low. Transient expression verified FcWD40-97's localization to the nucleus. Yeast two-hybrid (Y2H) and biomolecular fluorescence complementation (BiFC) assays revealed that FcWD40-97 interacts with FcMYB114, FcMYB123, and FcbHLH42 proteins in vitro and in vivo, showing that FcWD40-97 functions as a member of the MYB-bHLH-WD40 (MBW) complex in anthocyanin-biosynthesis regulation in fig. We therefore renamed FcWD40-97 as FcTTG1. Our results provide the first systematic analysis of the FcWD40 family and identification of FcTTG1 in fig pigmentation.
Keywords: MBW complex; TTG1; WD40; anthocyanin biosynthesis; expression profile; fig (Ficus carica L.).
Copyright © 2022 Fan, Zhai, Wang, Zhang, Song, Flaishman and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Fig (Ficus carica L.).Front Plant Sci. 2021 Oct 8;12:730692. doi: 10.3389/fpls.2021.730692. eCollection 2021. Front Plant Sci. 2021. PMID: 34691109 Free PMC article.
-
Differential color development and response to light deprivation of fig (Ficus carica L.) syconia peel and female flower tissues: transcriptome elucidation.BMC Plant Biol. 2019 May 23;19(1):217. doi: 10.1186/s12870-019-1816-9. BMC Plant Biol. 2019. PMID: 31122203 Free PMC article.
-
Anthocyanin accumulation is initiated by abscisic acid to enhance fruit color during fig (Ficus carica L.) ripening.J Plant Physiol. 2020 Aug;251:153192. doi: 10.1016/j.jplph.2020.153192. Epub 2020 Jun 2. J Plant Physiol. 2020. PMID: 32554070
-
Genome-wide in silico identification of glutathione S-transferase (GST) gene family members in fig (Ficus carica L.) and expression characteristics during fruit color development.PeerJ. 2023 Jan 25;11:e14406. doi: 10.7717/peerj.14406. eCollection 2023. PeerJ. 2023. PMID: 36718451 Free PMC article. Review.
-
GLABRA2, A Common Regulator for Epidermal Cell Fate Determination and Anthocyanin Biosynthesis in Arabidopsis.Int J Mol Sci. 2019 Oct 9;20(20):4997. doi: 10.3390/ijms20204997. Int J Mol Sci. 2019. PMID: 31601032 Free PMC article. Review.
Cited by
-
Genome-Wide Identification of the WD40 Gene Family in Walnut (Juglans regia L.) and Its Expression Profile in Different Colored Varieties.Int J Mol Sci. 2025 Jan 26;26(3):1071. doi: 10.3390/ijms26031071. Int J Mol Sci. 2025. PMID: 39940845 Free PMC article.
-
Comprehensive analysis of the Spartina alterniflora WD40 gene family reveals the regulatory role of SaTTG1 in plant development.Front Plant Sci. 2024 May 28;15:1390461. doi: 10.3389/fpls.2024.1390461. eCollection 2024. Front Plant Sci. 2024. PMID: 38863548 Free PMC article.
-
ERF100 regulated by ERF28 and NOR controls pectate lyase 7, modulating fig (Ficus carica L.) fruit softening.Plant Biotechnol J. 2025 Jul;23(7):2611-2626. doi: 10.1111/pbi.70085. Epub 2025 Apr 10. Plant Biotechnol J. 2025. PMID: 40209051 Free PMC article.
-
Integrated metabolomic and transcriptomic analysis of anthocyanin metabolism in wheat pericarp.BMC Genom Data. 2025 Jan 13;26(1):3. doi: 10.1186/s12863-024-01294-y. BMC Genom Data. 2025. PMID: 39806276 Free PMC article.
-
Genomic insights into the domestication and genetic basis of yield in papaya.Hortic Res. 2025 Feb 17;12(5):uhaf045. doi: 10.1093/hr/uhaf045. eCollection 2025 May. Hortic Res. 2025. PMID: 40236729 Free PMC article.
References
LinkOut - more resources
Full Text Sources

