Phylotranscriptomics Illuminates the Placement of Whole Genome Duplications and Gene Retention in Ferns
- PMID: 35909764
- PMCID: PMC9330400
- DOI: 10.3389/fpls.2022.882441
Phylotranscriptomics Illuminates the Placement of Whole Genome Duplications and Gene Retention in Ferns
Abstract
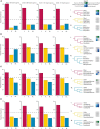
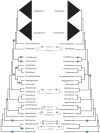
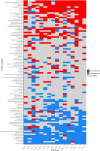
Ferns are the second largest clade of vascular plants with over 10,000 species, yet the generation of genomic resources for the group has lagged behind other major clades of plants. Transcriptomic data have proven to be a powerful tool to assess phylogenetic relationships, using thousands of markers that are largely conserved across the genome, and without the need to sequence entire genomes. We assembled the largest nuclear phylogenetic dataset for ferns to date, including 2884 single-copy nuclear loci from 247 transcriptomes (242 ferns, five outgroups), and investigated phylogenetic relationships across the fern tree, the placement of whole genome duplications (WGDs), and gene retention patterns following WGDs. We generated a well-supported phylogeny of ferns and identified several regions of the fern phylogeny that demonstrate high levels of gene tree-species tree conflict, which largely correspond to areas of the phylogeny that have been difficult to resolve. Using a combination of approaches, we identified 27 WGDs across the phylogeny, including 18 large-scale events (involving more than one sampled taxon) and nine small-scale events (involving only one sampled taxon). Most inferred WGDs occur within single lineages (e.g., orders, families) rather than on the backbone of the phylogeny, although two inferred events are shared by leptosporangiate ferns (excluding Osmundales) and Polypodiales (excluding Lindsaeineae and Saccolomatineae), clades which correspond to the majority of fern diversity. We further examined how retained duplicates following WGDs compared across independent events and found that functions of retained genes were largely convergent, with processes involved in binding, responses to stimuli, and certain organelles over-represented in paralogs while processes involved in transport, organelles derived from endosymbiotic events, and signaling were under-represented. To date, our study is the most comprehensive investigation of the nuclear fern phylogeny, though several avenues for future research remain unexplored.
Keywords: biased gene retention; fern; phylogenetics; polyploidy; transcriptome; whole genome duplication.
Copyright © 2022 Pelosi, Kim, Barbazuk and Sessa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Barker M. S., Kane N. C., Matvienko M., Kozik A., Michelmore R. W., Knapp S. J., et al. (2008). Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol. 25 2445–2455. 10.1093/molbev/msn187 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

