Identification of Six Genes as Diagnostic Markers for Colorectal Cancer Detection by Integrating Multiple Expression Profiles
- PMID: 35909904
- PMCID: PMC9337943
- DOI: 10.1155/2022/3850674
Identification of Six Genes as Diagnostic Markers for Colorectal Cancer Detection by Integrating Multiple Expression Profiles
Abstract
Background: Many studies have demonstrated the promising utility of DNA methylation and miRNA as biomarkers for colorectal cancer (CRC) early detection. However, mRNA is rarely reported. This study aimed to identify novel fecal-based mRNA signatures.
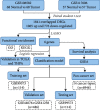
Methods: The differentially expressed genes (DEGs) were first determined between CRCs and matched normal samples by integrating multiple datasets. Then, Least Absolute Shrinkage and Selection Operator (LASSO) regression was used to reduce the number of candidates of aberrantly expressed genes. Next, the potential functions were investigated for the candidate signatures and their ability to detect CRC and pan-cancers was comprehensively evaluated.
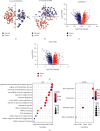
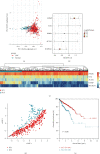
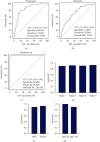
Results: We identified 1841 common DEGs in two independent datasets. Functional enrichment analysis revealed they were mainly related to extracellular structure, biosynthesis, and cell adhesion. The CRC classifier was established based on six genes screened by LASSO regression. Sensitivity, specificity, and area under the ROC curve (AUC) for CRC detection were 79.30%, 80.40%, and 0.85 (0.76-0.92) in the training set, and these indexes achieved 93.20%, 41.80%, and 0.73 (0.65-0.83) in the testing set. For validation set, the sensitivity, specificity, and AUC were 98.90%, 98.00%, and 0.97 (0.94-0.99). The average sensitivities exceeded 90.00% for CRCs with different clinical features. For adenomas detection, the sensitivity and specificity were 74.50% and 64.00%. Besides, the six genes obtained an average AUC of 0.855 for pan-cancer detection.
Conclusion: The six-gene signatures showed ability to detect CRC and pan-cancer samples, which could be served as potential diagnostic markers.
Copyright © 2022 Peijie Wu et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Machine Learning-Based Identification of Colon Cancer Candidate Diagnostics Genes.Biology (Basel). 2022 Feb 25;11(3):365. doi: 10.3390/biology11030365. Biology (Basel). 2022. PMID: 35336739 Free PMC article.
-
Identification and Validation of MicroRNA Profiles in Fecal Samples for Detection of Colorectal Cancer.Gastroenterology. 2020 Mar;158(4):947-957.e4. doi: 10.1053/j.gastro.2019.10.005. Epub 2019 Oct 14. Gastroenterology. 2020. PMID: 31622624
-
Fecal occult blood test for colorectal cancer screening: an evidence-based analysis.Ont Health Technol Assess Ser. 2009;9(10):1-40. Epub 2009 Sep 1. Ont Health Technol Assess Ser. 2009. PMID: 23074514 Free PMC article.
-
A systematic review and quantitative assessment of methylation biomarkers in fecal DNA and colorectal cancer and its precursor, colorectal adenoma.Mutat Res Rev Mutat Res. 2019 Jan-Mar;779:45-57. doi: 10.1016/j.mrrev.2019.01.003. Epub 2019 Jan 16. Mutat Res Rev Mutat Res. 2019. PMID: 31097151
-
Diagnostic value of aberrant gene methylation in stool samples for colorectal cancer or adenomas: a meta-analysis.Panminerva Med. 2015 Jun;57(2):55-64. Epub 2014 Sep 26. Panminerva Med. 2015. PMID: 25256821 Review.
Cited by
-
Nanoparticle-Based Drug Delivery Systems Targeting Tumor Microenvironment for Cancer Immunotherapy Resistance: Current Advances and Applications.Pharmaceutics. 2022 Sep 21;14(10):1990. doi: 10.3390/pharmaceutics14101990. Pharmaceutics. 2022. PMID: 36297426 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

