Studying Disease-Associated UBE3A Missense Variants Using Enhanced Sampling Molecular Simulations
- PMID: 35910155
- PMCID: PMC9330222
- DOI: 10.1021/acsomega.2c00959
Studying Disease-Associated UBE3A Missense Variants Using Enhanced Sampling Molecular Simulations
Abstract
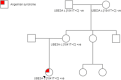
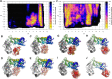
Missense variants in UBE3A underlie neurodevelopmental conditions such as Angelman Syndrome and Autism Spectrum Disorder, but the underlying molecular pathological consequences on protein folding and function are poorly understood. Here, we report a novel, maternally inherited, likely pathogenic missense variant in UBE3A (NM_000462.4(UBE3A_v001):(c.1841T>C) (p.(Leu614Pro))) in a child that presented with myoclonic epilepsy from 14 months, subsequent developmental regression from 16 months, and additional features consistent with Angelman Syndrome. To understand the impact of p.(Leu614Pro) on UBE3A, we used adiabatic biased molecular dynamics and metadynamics simulations to investigate conformational differences from wildtype proteins. Our results suggest that Leu614Pro substitution leads to less efficient binding and substrate processing compared to wildtype. Our results support the use of enhanced sampling molecular simulations to investigate the impact of missense UBE3A variants on protein function that underlies neurodevelopment and human disorders.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Richards S.; Aziz N.; Bale S.; Bick D.; Das S.; Gastier-Foster J.; Grody W. W.; Hegde M.; Lyon E.; Spector E.; et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. 10.1038/gim.2015.30. - DOI - PMC - PubMed
-
- Bossuyt S. N. V.; Punt A. M.; de Graaf I. J.; van den Burg J.; Williams M. G.; Heussler H.; Elgersma Y.; Distel B. Loss of nuclear UBE3A activity is the predominant cause of Angelman syndrome in individuals carrying UBE3A missense mutations. Hum. Mol. Genet. 2021, 30, 430–442. 10.1093/hmg/ddab050. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

