Genome-Wide Association Study of COVID-19 Outcomes Reveals Novel Host Genetic Risk Loci in the Serbian Population
- PMID: 35910207
- PMCID: PMC9329799
- DOI: 10.3389/fgene.2022.911010
Genome-Wide Association Study of COVID-19 Outcomes Reveals Novel Host Genetic Risk Loci in the Serbian Population
Abstract
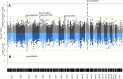
Host genetics, an important contributor to the COVID-19 clinical susceptibility and severity, currently is the focus of multiple genome-wide association studies (GWAS) in populations affected by the pandemic. This is the first study from Serbia that performed a GWAS of COVID-19 outcomes to identify genetic risk markers of disease severity. A group of 128 hospitalized COVID-19 patients from the Serbian population was enrolled in the study. We conducted a GWAS comparing (1) patients with pneumonia (n = 80) against patients without pneumonia (n = 48), and (2) severe (n = 34) against mild disease (n = 48) patients, using a genotyping array followed by imputation of missing genotypes. We have detected a significant signal associated with COVID-19 related pneumonia at locus 13q21.33, with a peak residing upstream of the gene KLHL1 (p = 1.91 × 10-8). Our study also replicated a previously reported COVID-19 risk locus at 3p21.31, identifying lead variants in SACM1L and LZTFL1 genes suggestively associated with pneumonia (p = 7.54 × 10-6) and severe COVID-19 (p = 6.88 × 10-7), respectively. Suggestive association with COVID-19 pneumonia has also been observed at chromosomes 5p15.33 (IRX, NDUFS6, MRPL36, p = 2.81 × 10-6), 5q11.2 (ESM1, p = 6.59 × 10-6), and 9p23 (TYRP1, LURAP1L, p = 8.69 × 10-6). The genes located in or near the risk loci are expressed in neural or lung tissues, and have been previously associated with respiratory diseases such as asthma and COVID-19 or reported as differentially expressed in COVID-19 gene expression profiling studies. Our results revealed novel risk loci for pneumonia and severe COVID-19 disease which could contribute to a better understanding of the COVID-19 host genetics in different populations.
Keywords: GWAS; SARS-CoV-2; genetic markers; pneumonia; severe disease.
Copyright © 2022 Zecevic, Kotur, Ristivojevic, Gasic, Skodric-Trifunovic, Stjepanovic, Stevanovic, Lavadinovic, Zukic, Pavlovic and Stankovic.
Conflict of interest statement
Author MZ was employed by the company Seven Bridges. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

