Structural Characterization of the Acer ukurunduense Chloroplast Genome Relative to Related Species in the Acer Genus
- PMID: 35910210
- PMCID: PMC9329572
- DOI: 10.3389/fgene.2022.849182
Structural Characterization of the Acer ukurunduense Chloroplast Genome Relative to Related Species in the Acer Genus
Abstract
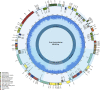
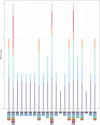
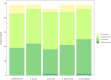
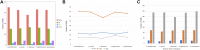
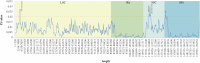
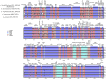
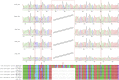
Acer ukurunduense refers to a deciduous tree distributed in Northeast Asia and is a widely used landscaping tree species. Although several studies have been conducted on the species' ecological and economic significance, limited information is available on its phylo-genomics. Our study newly constitutes the complete chloroplast genome of A. ukurunduense into a 156,645-bp circular DNA, which displayed a typical quadripartite structure. In addition, 133 genes were identified, containing 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes. In total, 107 simple sequence repeats and 49 repetitive sequences were observed. Thirty-two codons indicated that biased usages were estimated across 20 protein-coding genes (CDS) in A. ukurunduense. Four hotspot regions (trnK-UUU/rps16, ndhF/rpl32, rpl32/trnL-UAG, and ycf1) were detected among the five analyzed Acer species. Those hotspot regions may be useful molecular markers and contribute to future population genetics studies. The phylogenetic analysis demonstrated that A. ukurunduense is most closely associated with the species of Sect. Palmata. A. ukurunduense and A. pubipetiolatum var. pingpienense diverged in 22.11 Mya. We selected one of the hypervariable regions (trnK-UUU/rps16) to develop a new molecular marker and designed primers and confirmed that the molecular markers could accurately discriminate five Acer species through Sanger sequencing. By sequencing the cp genome of A. ukurunduense and comparing it with the relative species of Acer, we can effectively address the phylogenetic problems of Acer at the species level and provide insights into future research on population genetics and genetic diversity.
Keywords: Acer ukurunduense; Aceraceae; chloroplast genome; phylogenetic analysis; species identification.
Copyright © 2022 Ren, Liu, Yan, Jiang, Wang, Wang, Zhang, Liu, Sun, Gao and Ma.
Conflict of interest statement
WM was employed by Jiangsu Kanion Pharmaceutical Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Decoding the Chloroplast Genome of Korean endemic plant Acer okamotoanum: Comparative Genomics, Phylogenetic Insights, and Potential for Marker Development.Mol Biotechnol. 2025 Feb 5. doi: 10.1007/s12033-025-01383-y. Online ahead of print. Mol Biotechnol. 2025. PMID: 39907952
-
A systematic comparison of eight new plastome sequences from Ipomoea L.PeerJ. 2019 Mar 11;7:e6563. doi: 10.7717/peerj.6563. eCollection 2019. PeerJ. 2019. PMID: 30881765 Free PMC article.
-
The complete chloroplast genome of Acer Pubipetiolatum var. pingpienense (Sapindaceae).Mitochondrial DNA B Resour. 2022 Aug 8;7(8):1448-1450. doi: 10.1080/23802359.2022.2107443. eCollection 2022. Mitochondrial DNA B Resour. 2022. PMID: 35958062 Free PMC article.
-
The complete chloroplast genome sequence of the Korean maple tree (Acer pseudosieboldianum (Pax) Kom.).Mitochondrial DNA B Resour. 2023 Jun 25;8(6):691-694. doi: 10.1080/23802359.2023.2224623. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 37383607 Free PMC article.
-
Sequencing and Phylogenetic Analysis of the Chloroplast Genome of Three Apricot Species.Genes (Basel). 2023 Oct 18;14(10):1959. doi: 10.3390/genes14101959. Genes (Basel). 2023. PMID: 37895308 Free PMC article.
Cited by
-
Decoding the Chloroplast Genome of Korean endemic plant Acer okamotoanum: Comparative Genomics, Phylogenetic Insights, and Potential for Marker Development.Mol Biotechnol. 2025 Feb 5. doi: 10.1007/s12033-025-01383-y. Online ahead of print. Mol Biotechnol. 2025. PMID: 39907952
-
Comparative Genomic and Phylogenetic Analysis of Chloroplasts in Citrus paradisi Mac.cv. Cocktail.Genes (Basel). 2025 Apr 30;16(5):544. doi: 10.3390/genes16050544. Genes (Basel). 2025. PMID: 40428365 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

