QiShenYiQi Pill Ameliorates Cardiac Fibrosis After Pressure Overload-Induced Cardiac Hypertrophy by Regulating FHL2 and the Macrophage RP S19/TGF-β1 Signaling Pathway
- PMID: 35910357
- PMCID: PMC9326396
- DOI: 10.3389/fphar.2022.918335
QiShenYiQi Pill Ameliorates Cardiac Fibrosis After Pressure Overload-Induced Cardiac Hypertrophy by Regulating FHL2 and the Macrophage RP S19/TGF-β1 Signaling Pathway
Abstract
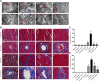
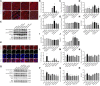
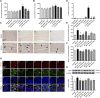
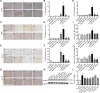
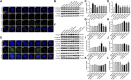
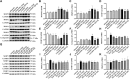
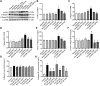
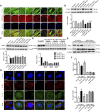
Purpose: Heart failure (HF) is a leading cause of morbidity and mortality worldwide, and it is characterized by cardiac hypertrophy and fibrosis. However, effective treatments are not available to block cardiac fibrosis after cardiac hypertrophy. The QiShenYiQi pill (QSYQ) is an effective treatment for chronic HF. However, the underlying mechanism remains unclear. Methods: In the present study, a pressure overload-induced cardiac hypertrophy model was established in rats by inducing ascending aortic stenosis for 4 weeks. QSYQ was administered for 6 weeks, and its effects on cardiac fibrosis, myocardial apoptosis, RP S19 release, macrophage polarization, TGF-β1 production, and TGF-β1/Smad signaling were analyzed. In vitro studies using H9C2, Raw264.7, and RDF cell models were performed to confirm the in vivo study findings and evaluate the contribution to the observed effects of the main ingredients of QSYQ, namely, astragaloside IV, notoginsenoside R1, 3,4-dihydroxyl-phenyl lactic acid, and Dalbergia odorifera T. C. Chen oil. The role of four-and-a-half LIM domains protein 2 (FHL2) in cardiac fibrosis and QSYQ's effects were assessed by small interfering RNAs (siRNAs). Results: QSYQ ameliorated cardiac fibrosis after pressure overload-induced cardiac hypertrophy and attenuated cardiomyocyte apoptosis, low FHL2 expression, and TGF-β1 release by the injured myocardium. QSYQ also inhibited the following: release of RP S19 from the injured myocardium, activation of C5a receptors in monocytes, polarization of macrophages, and release of TGF-β1. Moreover, QSYQ downregulated TGF-βR-II expression induced by TGF-β1 in fibroblasts and inhibited Smad protein activation and collagen release and deposition. Conclusion: The results showed that QSYQ inhibited myocardial fibrosis after pressure overload, which was mediated by RP S19-TGF-β1 signaling and decreased FHL2, thus providing support for QSYQ as a promising therapy for blocking myocardial fibrosis.
Keywords: FHL2; QiShenYiQi pill; RP S19; SMADs; TGF-β1; cardiac fibrosis; heart failure; pressure overload.
Copyright © 2022 Anwaier, Xie, Pan, Li, Yan, Wang, Chen, Weng, Sun, Chang, Fan, Han and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

