Identification and expression analysis of chlorophyll a/b binding protein gene family in grape (Vitis vinifera)
- PMID: 35910436
- PMCID: PMC9334500
- DOI: 10.1007/s12298-022-01204-5
Identification and expression analysis of chlorophyll a/b binding protein gene family in grape (Vitis vinifera)
Abstract
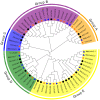
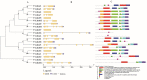
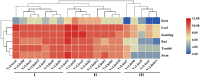
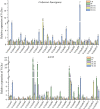
In higher plants, light capture of chlorophyll a/b binding protein (Lhc) plays a crucial role in the plant's response to adverse environment. So far, the family has not been systematically identified in grapes. In this study, 20 VvLhcs were identified in the grape genome, which were distributed in 13 of 19 grape chromosomes and divided into 7 developing branches. The results of gene duplication analysis showed that 6 VvLhcs formed fragment duplication events, while there was no tandem duplication in VvLhcs. Exon-intron structure analysis showed that they had a wide number of exons. Protein conserved motif analysis showed that VvLhcs contained more similar motif structures in the same phylogenetic branch. The cis-acting elements in the VvLhcs promoter region mainly respond to light, plant hormones and abiotic stresses. In addition, qRT-PCR results showed that different proportions of salt stress and red-blue light affected the expression of VvLhcs and the expression patterns of genes in different grape varieties were different. The results for further study on different grape varieties in different combinations of red and blue light of the Lhc provide a theoretical basis.
Supplementary information: The online version contains supplementary material available at 10.1007/s12298-022-01204-5.
Keywords: Gene family identification; Grape; Lhc; Red-blue light; Salt stress.
© Prof. H.S. Srivastava Foundation for Science and Society 2022.
Conflict of interest statement
Conflict of interestThe authors declare no conflict of interest.
Figures


Similar articles
-
Genome-wide identification and expression analysis of PIN gene family under phytohormone and abiotic stresses in Vitis Vinifera L.Physiol Mol Biol Plants. 2022 Oct;28(10):1905-1919. doi: 10.1007/s12298-022-01239-8. Epub 2022 Nov 5. Physiol Mol Biol Plants. 2022. PMID: 36484025 Free PMC article.
-
The metacaspase gene family of Vitis vinifera L.: characterization and differential expression during ovule abortion in stenospermocarpic seedless grapes.Gene. 2013 Oct 10;528(2):267-76. doi: 10.1016/j.gene.2013.06.062. Epub 2013 Jul 9. Gene. 2013. PMID: 23845786
-
Genome-Wide Identification and Expression Analysis of GA2ox, GA3ox, and GA20ox Are Related to Gibberellin Oxidase Genes in Grape (VitisVinifera L.).Genes (Basel). 2019 Sep 5;10(9):680. doi: 10.3390/genes10090680. Genes (Basel). 2019. PMID: 31492001 Free PMC article.
-
Genome-wide identification of small heat-shock protein (HSP20) gene family in grape and expression profile during berry development.BMC Plant Biol. 2019 Oct 17;19(1):433. doi: 10.1186/s12870-019-2031-4. BMC Plant Biol. 2019. PMID: 31623556 Free PMC article.
-
Genome-wide identification and characterization of the NF-Y gene family in grape (vitis vinifera L.).BMC Genomics. 2016 Aug 11;17(1):605. doi: 10.1186/s12864-016-2989-3. BMC Genomics. 2016. PMID: 27516172 Free PMC article.
Cited by
-
Identification of the Light-Harvesting Chlorophyll a/b Binding Protein Gene Family in Peach (Prunus persica L.) and Their Expression under Drought Stress.Genes (Basel). 2023 Jul 19;14(7):1475. doi: 10.3390/genes14071475. Genes (Basel). 2023. PMID: 37510379 Free PMC article.
-
Chlorophyll a/b-binding protein gene AcLhcb1.1 promotes chlorophyll accumulation and improves cold stress resistance in kiwifruit.BMC Plant Biol. 2025 Aug 27;25(1):1142. doi: 10.1186/s12870-025-07152-y. BMC Plant Biol. 2025. PMID: 40866862 Free PMC article.
-
Shading Treatment Reduces Grape Sugar Content by Suppressing Photosynthesis-Antenna Protein Pathway Gene Expression in Grape Berries.Int J Mol Sci. 2024 May 5;25(9):5029. doi: 10.3390/ijms25095029. Int J Mol Sci. 2024. PMID: 38732247 Free PMC article.
-
Chlorophyllase (PsCLH1) and light-harvesting chlorophyll a/b binding protein 1 (PsLhcb1) and PsLhcb5 maintain petal greenness in Paeonia suffruticosa 'Lv Mu Yin Yu'.J Adv Res. 2025 Jul;73:173-185. doi: 10.1016/j.jare.2024.09.003. Epub 2024 Sep 3. J Adv Res. 2025. PMID: 39236974 Free PMC article.
-
The transcriptional analysis of pepper shed light on a proviral role of light-harvesting chlorophyll a/b binding protein 13 during infection of pepper mild mottle virus.Front Plant Sci. 2025 Jan 27;16:1533151. doi: 10.3389/fpls.2025.1533151. eCollection 2025. Front Plant Sci. 2025. PMID: 39931497 Free PMC article.
References
-
- Broglie R, Bellemare G, Bartlett SG, Chua NH, Cashmore AR. Cloned DNA sequences complementary to mRNAs encoding precursors to the small subunit of ribulose-1,5-bisphosphate carboxylase and a chlorophyll a/b binding polypeptide. Proc Natl Acad Sci USA. 1981;78:7304–7308. doi: 10.1073/pnas.78.12.7304. - DOI - PMC - PubMed
-
- Brunner H, Rüdiger W. The greening process in cress seedlings IV. Light regulated expression of single Lhc genes. J Photochem Photobiol B Biol. 1995;27:257–263. doi: 10.1016/1011-1344(94)07076-Z. - DOI
LinkOut - more resources
Full Text Sources
