LARGE-SCALE MULTIPLE INFERENCE OF COLLECTIVE DEPENDENCE WITH APPLICATIONS TO PROTEIN FUNCTION
- PMID: 35910493
- PMCID: PMC9337751
- DOI: 10.1214/20-aoas1431
LARGE-SCALE MULTIPLE INFERENCE OF COLLECTIVE DEPENDENCE WITH APPLICATIONS TO PROTEIN FUNCTION
Abstract
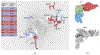
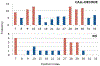
Measuring the dependence of k ≥ 3 random variables and drawing inference from such higher-order dependences are scientifically important yet challenging. Motivated here by protein coevolution with multivariate categorical features, we consider an information theoretic measure of higher-order dependence. The proposed collective dependence is a symmetrization of differential interaction information which generalizes the mutual information of a pair of random variables. We show that the collective dependence can be easily estimated and facilitates a test on the dependence of k ≥ 3 random variables. Upon carefully exploring the null space of collective dependence, we devise a Classification-Assisted Large scaLe inference procedure to DEtect significant k-COllective DEpendence among d ≥ k random variables, with the false discovery rate controlled. Finite sample performance of our method is examined via simulations. We apply this method to the multiple protein sequence alignment data to study the residue or position coevolution for two protein families, the elongation factor P family and the zinc knuckle family. We identify novel functional triplets of amino acid residues, whose contributions to the protein function are further investigated. These confirm that the collective dependence does yield additional information important for understanding the protein coevolution compared to the pairwise measures.
Keywords: Collective dependence; false discovery rate; information theoretic measure; multiple testing; protein coevolution; structural biology.
Figures




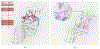
Similar articles
-
Sequence coevolution between RNA and protein characterized by mutual information between residue triplets.PLoS One. 2012;7(1):e30022. doi: 10.1371/journal.pone.0030022. Epub 2012 Jan 18. PLoS One. 2012. PMID: 22279560 Free PMC article.
-
Multiple Testing of Submatrices of a Precision Matrix with Applications to Identification of Between Pathway Interactions.J Am Stat Assoc. 2018;113(521):328-339. doi: 10.1080/01621459.2016.1251930. Epub 2017 Sep 26. J Am Stat Assoc. 2018. PMID: 29881130 Free PMC article.
-
Genotype distribution-based inference of collective effects in genome-wide association studies: insights to age-related macular degeneration disease mechanism.BMC Genomics. 2016 Aug 30;17(1):695. doi: 10.1186/s12864-016-2871-3. BMC Genomics. 2016. PMID: 27576376 Free PMC article.
-
The role of coevolutionary signatures in protein interaction dynamics, complex inference, molecular recognition, and mutational landscapes.Curr Opin Struct Biol. 2019 Jun;56:179-186. doi: 10.1016/j.sbi.2019.03.024. Epub 2019 Apr 28. Curr Opin Struct Biol. 2019. PMID: 31029927 Review.
-
Inferring the roles of individuals in collective systems using information-theoretic measures of influence.Biophys Physicobiol. 2024 Mar 22;21(Supplemental):e211014. doi: 10.2142/biophysico.bppb-v21.s014. eCollection 2024. Biophys Physicobiol. 2024. PMID: 39175852 Free PMC article. Review.
Cited by
-
General strategies for using amino acid sequence data to guide biochemical investigation of protein function.Biochem Soc Trans. 2022 Dec 16;50(6):1847-1858. doi: 10.1042/BST20220849. Biochem Soc Trans. 2022. PMID: 36416676 Free PMC article. Review.
References
-
- Basharin GP (1959). On a statistical estimate for the entropy of a sequence of independent random variables. Theory Probab. Appl 4 333–336. MR0127457 10.1137/1104033 - DOI
-
- Bell AJ (2003). The co-information lattices. In Proceedings of the Fifth International Workshop on Independent Component Analysis and Blind Signal Separation: IC 2003.
-
- Benjamini Y and Hochberg Y (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B 57 289–300. MR1325392
Grants and funding
LinkOut - more resources
Full Text Sources
