Forest tree species distribution for Europe 2000-2020: mapping potential and realized distributions using spatiotemporal machine learning
- PMID: 35910765
- PMCID: PMC9332400
- DOI: 10.7717/peerj.13728
Forest tree species distribution for Europe 2000-2020: mapping potential and realized distributions using spatiotemporal machine learning
Abstract
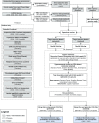
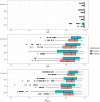
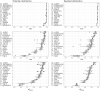
This article describes a data-driven framework based on spatiotemporal machine learning to produce distribution maps for 16 tree species (Abies alba Mill., Castanea sativa Mill., Corylus avellana L., Fagus sylvatica L., Olea europaea L., Picea abies L. H. Karst., Pinus halepensis Mill., Pinus nigra J. F. Arnold, Pinus pinea L., Pinus sylvestris L., Prunus avium L., Quercus cerris L., Quercus ilex L., Quercus robur L., Quercus suber L. and Salix caprea L.) at high spatial resolution (30 m). Tree occurrence data for a total of three million of points was used to train different algorithms: random forest, gradient-boosted trees, generalized linear models, k-nearest neighbors, CART and an artificial neural network. A stack of 305 coarse and high resolution covariates representing spectral reflectance, different biophysical conditions and biotic competition was used as predictors for realized distributions, while potential distribution was modelled with environmental predictors only. Logloss and computing time were used to select the three best algorithms to tune and train an ensemble model based on stacking with a logistic regressor as a meta-learner. An ensemble model was trained for each species: probability and model uncertainty maps of realized distribution were produced for each species using a time window of 4 years for a total of six distribution maps per species, while for potential distributions only one map per species was produced. Results of spatial cross validation show that the ensemble model consistently outperformed or performed as good as the best individual model in both potential and realized distribution tasks, with potential distribution models achieving higher predictive performances (TSS = 0.898, R2 logloss = 0.857) than realized distribution ones on average (TSS = 0.874, R2 logloss = 0.839). Ensemble models for Q. suber achieved the best performances in both potential (TSS = 0.968, R2 logloss = 0.952) and realized (TSS = 0.959, R2 logloss = 0.949) distribution, while P. sylvestris (TSS = 0.731, 0.785, R2 logloss = 0.585, 0.670, respectively, for potential and realized distribution) and P. nigra (TSS = 0.658, 0.686, R2 logloss = 0.623, 0.664) achieved the worst. Importance of predictor variables differed across species and models, with the green band for summer and the Normalized Difference Vegetation Index (NDVI) for fall for realized distribution and the diffuse irradiation and precipitation of the driest quarter (BIO17) being the most frequent and important for potential distribution. On average, fine-resolution models outperformed coarse resolution models (250 m) for realized distribution (TSS = +6.5%, R2 logloss = +7.5%). The framework shows how combining continuous and consistent Earth Observation time series data with state of the art machine learning can be used to derive dynamic distribution maps. The produced predictions can be used to quantify temporal trends of potential forest degradation and species composition change.
Keywords: Ecological niche; Ensemble modeling; High resolution; Imbalanced data; Machine learning; Presence-absence; Spatiotemporal modeling; Species distribution model; Stacked generalization; Tree species.
© 2022 Bonannella et al.
Conflict of interest statement
The authors declare that they have no competing interests. Carmelo Bonannella, Tomislav Hengl and Leandro Parente declare that they are officially employed by OpenGeoHub.
Figures





References
-
- Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, Anderson RP. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography. 2015;38(5):541–545. doi: 10.1111/ecog.01132. - DOI
-
- Anand A, Pandey MK, Srivastava PK, Gupta A, Khan ML. Integrating multi-sensors data for species distribution mapping using deep learning and envelope models. Remote Sensing. 2021;13(16):3284. doi: 10.3390/rs13163284. - DOI
-
- Andrewartha HG, Birch LC. The distribution and abundance of animals. Chicago: The University of Chicago Press; 1954.
-
- Astola H, Häme T, Sirro L, Molinier M, Kilpi J. Comparison of Sentinel-2 and Landsat 8 imagery for forest variable prediction in boreal region. Remote Sensing of Environment. 2019;223(1):257–273. doi: 10.1016/j.rse.2019.01.019. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

