Distinct Type I Interferon Subtypes Differentially Stimulate T Cell Responses in HIV-1-Infected Individuals
- PMID: 35911692
- PMCID: PMC9326074
- DOI: 10.3389/fimmu.2022.936918
Distinct Type I Interferon Subtypes Differentially Stimulate T Cell Responses in HIV-1-Infected Individuals
Abstract
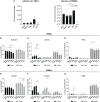
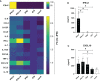
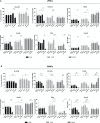
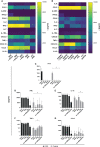
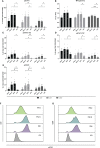
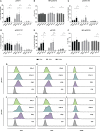
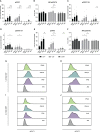
The expression of type I interferons (IFNs) is one of the immediate host responses during most viral infections. The type I IFN family consists of numerous highly conserved IFNα subtypes, IFNβ, and some others. Although these IFNα subtypes were initially believed to act interchangeably, their discrete biological properties are nowadays widely accepted. Subtype-specific antiviral, immunomodulatory, and anti-proliferative activities were reported explained by differences in receptor affinity, downstream signaling events, and individual IFN-stimulated gene expression patterns. Type I IFNs and increased IFN signatures potentially linked to hyperimmune activation of T cells are critically discussed for chronic HIV (human immunodeficiency virus) infection. Here, we aimed to analyze the broad immunological effects of specific type I IFN subtypes (IFNα2, IFNα14, and IFNβ) on T and NK cell subsets during HIV-1 infection in vitro and ex vivo. Stimulation with IFNα14 and IFNβ significantly increased frequencies of degranulating (CD107a+) gut-derived CD4+ T cells and blood-derived T and NK cells. However, frequencies of IFNγ-expressing T cells were strongly reduced after stimulation with IFNα14 and IFNβ. Phosphorylation of downstream molecules was not only IFN subtype-specific; also, significant differences in STAT5 phosphorylation were observed in both healthy peripheral blood mononuclear cells (PBMCs) and PBMCs of HIV-infected individuals, but this effect was less pronounced in healthy gut-derived lamina propria mononuclear cells (LPMCs), assuming cell and tissue specific discrepancies. In conclusion, we observed distinct type I IFN subtype-specific potencies in stimulating T and NK cell responses during HIV-1-infection.
Keywords: CD4+ T cells; CD8+ T cells; HIV; LPMCs; NK cells; type I IFNs.
Copyright © 2022 Karakoese, Schwerdtfeger, Karsten, Esser, Dittmer and Sutter.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
IFNα Subtypes in HIV Infection and Immunity.Viruses. 2024 Feb 27;16(3):364. doi: 10.3390/v16030364. Viruses. 2024. PMID: 38543729 Free PMC article. Review.
-
Qualitative Differences Between the IFNα subtypes and IFNβ Influence Chronic Mucosal HIV-1 Pathogenesis.PLoS Pathog. 2020 Oct 16;16(10):e1008986. doi: 10.1371/journal.ppat.1008986. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33064743 Free PMC article.
-
Interferon-α Subtypes in an Ex Vivo Model of Acute HIV-1 Infection: Expression, Potency and Effector Mechanisms.PLoS Pathog. 2015 Nov 3;11(11):e1005254. doi: 10.1371/journal.ppat.1005254. eCollection 2015. PLoS Pathog. 2015. PMID: 26529416 Free PMC article.
-
Type I interferon upregulates Bak and contributes to T cell loss during human immunodeficiency virus (HIV) infection.PLoS Pathog. 2013;9(10):e1003658. doi: 10.1371/journal.ppat.1003658. Epub 2013 Oct 10. PLoS Pathog. 2013. PMID: 24130482 Free PMC article. Clinical Trial.
-
Interferon α subtypes in HIV infection.Cytokine Growth Factor Rev. 2018 Apr;40:13-18. doi: 10.1016/j.cytogfr.2018.02.002. Epub 2018 Feb 13. Cytokine Growth Factor Rev. 2018. PMID: 29475588 Review.
Cited by
-
IFNα Subtypes in HIV Infection and Immunity.Viruses. 2024 Feb 27;16(3):364. doi: 10.3390/v16030364. Viruses. 2024. PMID: 38543729 Free PMC article. Review.
-
Sex-dependent differences in type I IFN-induced natural killer cell activation.Front Immunol. 2023 Dec 15;14:1277967. doi: 10.3389/fimmu.2023.1277967. eCollection 2023. Front Immunol. 2023. PMID: 38162640 Free PMC article.
-
Immunotherapy-induced cytotoxic T follicular helper cells reduce numbers of retrovirus-infected reservoir cells in B cell follicles.PLoS Pathog. 2023 Oct 26;19(10):e1011725. doi: 10.1371/journal.ppat.1011725. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37883584 Free PMC article.
-
Type I interferon subtypes differentially activate the anti-leukaemic function of natural killer cells.Front Immunol. 2022 Nov 24;13:1050718. doi: 10.3389/fimmu.2022.1050718. eCollection 2022. Front Immunol. 2022. PMID: 36505400 Free PMC article.
-
IFNα subtype-specific susceptibility of HBV in the course of chronic infection.Front Immunol. 2022 Oct 14;13:1017753. doi: 10.3389/fimmu.2022.1017753. eCollection 2022. Front Immunol. 2022. PMID: 36311794 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

