Susceptibility and Severity of COVID-19 Are Both Associated With Lower Overall Viral-Peptide Binding Repertoire of HLA Class I Molecules, Especially in Younger People
- PMID: 35911710
- PMCID: PMC9331187
- DOI: 10.3389/fimmu.2022.891816
Susceptibility and Severity of COVID-19 Are Both Associated With Lower Overall Viral-Peptide Binding Repertoire of HLA Class I Molecules, Especially in Younger People
Abstract
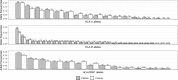
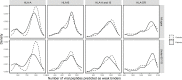
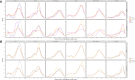
An important number of studies have been conducted on the potential association between human leukocyte antigen (HLA) genes and COVID-19 susceptibility and severity since the beginning of the pandemic. However, case-control and peptide-binding prediction methods tended to provide inconsistent conclusions on risk and protective HLA alleles, whereas some researchers suggested the importance of considering the overall capacity of an individual's HLA Class I molecules to present SARS-CoV-2-derived peptides. To close the gap between these approaches, we explored the distributions of HLA-A, -B, -C, and -DRB1 1st-field alleles in 142 Iranian patients with COVID-19 and 143 ethnically matched healthy controls, and applied in silico predictions of bound viral peptides for each individual's HLA molecules. Frequency comparison revealed the possible predisposing roles of HLA-A*03, B*35, and DRB1*16 alleles and the protective effect of HLA-A*32, B*58, B*55, and DRB1*14 alleles in the viral infection. None of these results remained significant after multiple testing corrections, except HLA-A*03, and no allele was associated with severity, either. Compared to peptide repertoires of individual HLA molecules that are more likely population-specific, the overall coverage of virus-derived peptides by one's HLA Class I molecules seemed to be a more prominent factor associated with both COVID-19 susceptibility and severity, which was independent of affinity index and threshold chosen, especially for people under 60 years old. Our results highlight the effect of the binding capacity of different HLA Class I molecules as a whole, and the more essential role of HLA-A compared to HLA-B and -C genes in immune responses against SARS-CoV-2 infection.
Keywords: COVID-19; HLA; HLA binding prediction; SARS-CoV-2-derived peptides; overall binding repertoire.
Copyright © 2022 Basir, Majzoobi, Ebrahimi, Noroozbeygi, Hashemi, Keramat, Mamani, Eini, Alizadeh, Solgi and Di.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Possible role of HLA class-I genotype in SARS-CoV-2 infection and progression: A pilot study in a cohort of Covid-19 Spanish patients.Clin Immunol. 2020 Oct;219:108572. doi: 10.1016/j.clim.2020.108572. Epub 2020 Aug 15. Clin Immunol. 2020. PMID: 32810602 Free PMC article.
-
HLA-C*04:01 Affects HLA Class I Heterozygosity and Predicted Affinity to SARS-CoV-2 Peptides, and in Combination With Age and Sex of Armenian Patients Contributes to COVID-19 Severity.Front Immunol. 2022 Feb 3;13:769900. doi: 10.3389/fimmu.2022.769900. eCollection 2022. Front Immunol. 2022. PMID: 35185875 Free PMC article.
-
Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2.J Virol. 2020 Jun 16;94(13):e00510-20. doi: 10.1128/JVI.00510-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32303592 Free PMC article.
-
HLA variation and antigen presentation in COVID-19 and SARS-CoV-2 infection.Curr Opin Immunol. 2022 Jun;76:102178. doi: 10.1016/j.coi.2022.102178. Epub 2022 Mar 25. Curr Opin Immunol. 2022. PMID: 35462277 Free PMC article. Review.
-
HLA alleles associated with COVID-19 susceptibility and severity in different populations: a systematic review.Egypt J Med Hum Genet. 2023;24(1):10. doi: 10.1186/s43042-023-00390-5. Epub 2023 Jan 22. Egypt J Med Hum Genet. 2023. PMID: 36710951 Free PMC article. Review.
Cited by
-
The HLA-B -21 M/T dimorphism associates with disease severity in COVID-19.Genes Immun. 2025 Feb;26(1):70-74. doi: 10.1038/s41435-024-00302-6. Epub 2024 Nov 1. Genes Immun. 2025. PMID: 39487235 Free PMC article.
-
Association of Alleles of Human Leukocyte Antigen Class II Genes and Severity of COVID-19 in Patients of the 'Red Zone' of the Endocrinology Research Center, Moscow, Russia.Diseases. 2022 Nov 2;10(4):99. doi: 10.3390/diseases10040099. Diseases. 2022. PMID: 36412593 Free PMC article.
-
The demographic, laboratory and genetic factors associated with long Covid-19 syndrome: a case-control study.Clin Exp Med. 2024 Jan 17;24(1):1. doi: 10.1007/s10238-023-01256-1. Clin Exp Med. 2024. PMID: 38231284 Free PMC article.
-
HLA Polymorphisms and COVID-19 Susceptibility and Severity: Insights From an Iranian Patients Cohort.J Cell Mol Med. 2025 May;29(9):e70570. doi: 10.1111/jcmm.70570. J Cell Mol Med. 2025. PMID: 40366340 Free PMC article.
-
Influence of HLA Class I and II Polymorphisms on COVID-19 Severity in a South Brazilian Population.Int J Mol Sci. 2025 Jun 2;26(11):5341. doi: 10.3390/ijms26115341. Int J Mol Sci. 2025. PMID: 40508150 Free PMC article.
References
-
- Who Coronavirus Dashboard . Geneva: World Health Organization (2022). Available at: https://covid19.who.int/ (Accessed 2022).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

