Differential Impacts of HHV-6A versus HHV-6B Infection in Differentiated Human Neural Stem Cells
- PMID: 35911725
- PMCID: PMC9326508
- DOI: 10.3389/fimmu.2022.847106
Differential Impacts of HHV-6A versus HHV-6B Infection in Differentiated Human Neural Stem Cells
Abstract
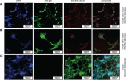
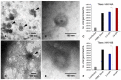
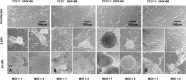
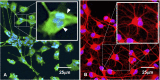
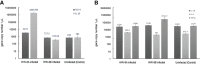
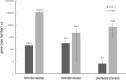
Within the family Herpesviridae, sub-family β-herpesvirinae, and genus Roseolovirus, there are only three human herpesviruses that have been described: HHV-6A, HHV-6B, and HHV-7. Initially, HHV-6A and HHV-6B were considered as two variants of the same virus (i.e., HHV6). Despite high overall genetic sequence identity (~90%), HHV-6A and HHV-6B are now recognized as two distinct viruses. Sequence divergence (e.g., >30%) in key coding regions and significant differences in physiological and biochemical profiles (e.g., use of different receptors for viral entry) underscore the conclusion that HHV-6A and HHV-6B are distinct viruses of the β-herpesvirinae. Despite these viruses being implicated as causative agents in several nervous system disorders (e.g., multiple sclerosis, epilepsy, and chronic fatigue syndrome), the mechanisms of action and relative contributions of each virus to neurological dysfunction are unclear. Unresolved questions regarding differences in cell tropism, receptor use and binding affinity (i.e., CD46 versus CD134), host neuro-immunological responses, and relative virulence between HHV-6A versus HHV-6B prevent a complete characterization. Although it has been shown that both HHV-6A and HHV-6B can infect glia (and, recently, cerebellar Purkinje cells), cell tropism of HHV-6A versus HHV-6B for different nerve cell types remains vague. In this study, we show that both viruses can infect different nerve cell types (i.e., glia versus neurons) and different neurotransmitter phenotypes derived from differentiated human neural stem cells. As demonstrated by immunofluorescence, HHV-6A and HHV-6B productively infect VGluT1-containing cells (i.e., glutamatergic neurons) and dopamine-containing cells (i.e., dopaminergic neurons). However, neither virus appears to infect GAD67-containing cells (i.e., GABAergic neurons). As determined by qPCR, expression of immunological factors (e.g., cytokines) in cells infected with HHV-6A versus HHV6-B also differs. These data along with morphometric and image analyses of infected differentiated neural stem cell cultures indicate that while HHV-6B may have greater opportunity for transmission, HHV-6A induces more severe cytopathic effects (e.g., syncytia) at the same post-infection end points. Cumulatively, results suggest that HHV-6A is more virulent than HHV-6B in susceptible cells, while neither virus productively infects GABAergic cells. Consistency between these in vitro data and in vivo experiments would provide new insights into potential mechanisms for HHV6-induced epileptogenesis.
Keywords: cell tropism; epilepsy; human herpesvirus 6; immunological response; neural stem cells; roseolovirus.
Copyright © 2022 Bahramian, Furr, Wu and Ceballos.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

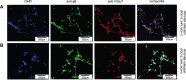
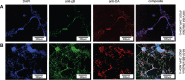



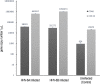
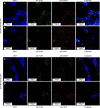
Similar articles
-
A Murine Herpesvirus Closely Related to Ubiquitous Human Herpesviruses Causes T-Cell Depletion.J Virol. 2017 Apr 13;91(9):e02463-16. doi: 10.1128/JVI.02463-16. Print 2017 May 1. J Virol. 2017. PMID: 28179532 Free PMC article.
-
Immunomodulation and immunosuppression by human herpesvirus 6A and 6B.Future Virol. 2013 Mar;8(3):273-287. doi: 10.2217/fvl.13.7. Future Virol. 2013. PMID: 24163703 Free PMC article.
-
gp96 Is Critical for both Human Herpesvirus 6A (HHV-6A) and HHV-6B Infections.J Virol. 2020 Jun 16;94(13):e00311-20. doi: 10.1128/JVI.00311-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295911 Free PMC article.
-
The role of herpesvirus 6A and 6B in multiple sclerosis and epilepsy.Scand J Immunol. 2020 Dec;92(6):e12984. doi: 10.1111/sji.12984. Epub 2020 Oct 23. Scand J Immunol. 2020. PMID: 33037649 Free PMC article. Review.
-
Recent topics related to human herpesvirus 6 cell tropism.Cell Microbiol. 2009 Jul;11(7):1001-6. doi: 10.1111/j.1462-5822.2009.01312.x. Epub 2009 Mar 12. Cell Microbiol. 2009. PMID: 19290911 Review.
Cited by
-
Human Herpesvirus-6B Infection and Alterations of Gut Microbiome in Patients with Fibromyalgia: A Pilot Study.Biomolecules. 2024 Oct 12;14(10):1291. doi: 10.3390/biom14101291. Biomolecules. 2024. PMID: 39456224 Free PMC article.
-
Viruses and the Brain-A Relationship Prone to Trouble.Viruses. 2025 Jan 31;17(2):203. doi: 10.3390/v17020203. Viruses. 2025. PMID: 40006958 Free PMC article. Review.
-
The Association of HHV-6 and the TNF-α (-308G/A) Promotor with Major Depressive Disorder Patients and Healthy Controls in Thailand.Viruses. 2023 Sep 8;15(9):1898. doi: 10.3390/v15091898. Viruses. 2023. PMID: 37766304 Free PMC article.
-
Child With Human Herpesvirus 6 and Bell's Palsy: Case Report and Overview of the Literature.Cureus. 2025 May 21;17(5):e84563. doi: 10.7759/cureus.84563. eCollection 2025 May. Cureus. 2025. PMID: 40546589 Free PMC article.
-
Human Herpesvirus 6A Infection-Associated Acute Anterior Uveitis.J Inflamm Res. 2024 Dec 24;17:11577-11585. doi: 10.2147/JIR.S489178. eCollection 2024. J Inflamm Res. 2024. PMID: 39735893 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

