Antibody-Antigen Binding Interface Analysis in the Big Data Era
- PMID: 35911958
- PMCID: PMC9329859
- DOI: 10.3389/fmolb.2022.945808
Antibody-Antigen Binding Interface Analysis in the Big Data Era
Abstract
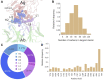
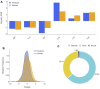
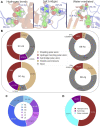
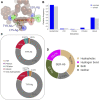
Antibodies have become the Swiss Army tool for molecular biology and nanotechnology. Their outstanding ability to specifically recognise molecular antigens allows their use in many different applications from medicine to the industry. Moreover, the improvement of conventional structural biology techniques (e.g., X-ray, NMR) as well as the emergence of new ones (e.g., Cryo-EM), have permitted in the last years a notable increase of resolved antibody-antigen structures. This offers a unique opportunity to perform an exhaustive structural analysis of antibody-antigen interfaces by employing the large amount of data available nowadays. To leverage this factor, different geometric as well as chemical descriptors were evaluated to perform a comprehensive characterization.
Keywords: antibody-antigen complex; complementarity-determining region (CDR); epitope analysis; paratope analysis; structure analysis and characterization.
Copyright © 2022 Reis, Barletta, Gagliardi, Fortuna, Soler and Rocchia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Cignoni P., Callieri M., Corsini M., Dellepiane M., Ganovelli F., Ranzuglia G. (2008). “MeshLab: an Open-Source Mesh Processing Tool,” in Eurographics Italian Chapter Conference. Editors Scarano V., Chiara R. D., Erra U. (Champagne-Ardenne: The Eurographics Association; ). 10.2312/LocalChapterEvents/ItalChap/ItalianChapConf2008/129-136 - DOI
-
- [Dataset] Muntoni A., Cignoni P. (2021). PyMeshLab. 10.5281/zenodo.4438750 - DOI
-
- Decherchi S., Colmenares J., Catalano C. E., Spagnuolo M., Alexov E., Rocchia W. (2013). Between Algorithm and Model: Different Molecular Surface Definitions for the Poisson-Boltzmann Based Electrostatic Characterization of Biomolecules in Solution. Commun. Comput. Phys. 13, 61–89. 10.4208/cicp.050711.111111s - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

