Necroptosis-Related Genes Signatures Identified Molecular Subtypes and Underlying Mechanisms in Hepatocellular Carcinoma
- PMID: 35912224
- PMCID: PMC9326098
- DOI: 10.3389/fonc.2022.875264
Necroptosis-Related Genes Signatures Identified Molecular Subtypes and Underlying Mechanisms in Hepatocellular Carcinoma
Abstract
Background: Although emerging evidence supports the relationship between necroptosis (NEC) related genes and hepatocellular carcinoma (HCC), the contribution of these necroptosis-related genes to the development, prognosis, and immunotherapy of HCC is unclear.
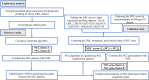
Methods: The expression of genes and relevant clinical information were downloaded from TCGA-LIHC, LIRI-JP, GSE14520/NCI, GSE36376, GSE76427, GSE20140, GSE27150, and IMvigor210 datasets. Next, we used an unsupervised clustering method to assign the samples into phenotype clusters base on 15 necroptosis-related genes. Subsequently, we constructed a NEC score based on NEC phenotype-related prognostic genes to quantify the necroptosis related subtypes of individual patients.
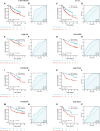
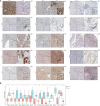
Results: We divided the samples into the high and low NEC score groups, and the high NEC score showed a poor prognosis. Simultaneously, NEC score is an effective and stable model and had a good performance in predicting the prognosis of HCC patients. A high NEC score was characterized by activation of the stroma and increased levels of immune infiltration. A high NEC score was also related to low expression of immune checkpoint molecules (PD-1/PD-L1). Importantly, the established NEC score would contribute to predicting the response to anti-PD-1/L1 immunotherapy.
Conclusions: Our study provide a comprehensive analysis of necroptosis-related genes in HCC. Stratification based on the NEC score may enable HCC patients to benefit more from immunotherapy and help identify new cancer treatment strategies.
Keywords: data mining; hepatocellular carcinoma; immunotherapy; necroptosis; tumor microenvironment.
Copyright © 2022 Wei, Hou, Li, Yao, Wang, Zheng, Mo, Chen and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Identification and characterization of a novel molecular classification based on disulfidptosis-related genes to predict prognosis and immunotherapy efficacy in hepatocellular carcinoma.Aging (Albany NY). 2023 Jul 3;15(13):6135-6151. doi: 10.18632/aging.204809. Epub 2023 Jul 3. Aging (Albany NY). 2023. PMID: 37399661 Free PMC article.
-
Oncogenic signaling pathway mediated by Notch pathway-related genes induces immunosuppression and immunotherapy resistance in hepatocellular carcinoma.Immunogenetics. 2022 Dec;74(6):539-557. doi: 10.1007/s00251-022-01273-6. Epub 2022 Jul 27. Immunogenetics. 2022. PMID: 35895154
-
Prognosis Risk Model Based on Necroptosis-Related Signature for Bladder Cancer.Genes (Basel). 2022 Nov 15;13(11):2120. doi: 10.3390/genes13112120. Genes (Basel). 2022. PMID: 36421795 Free PMC article.
-
Identification and analysis of necroptosis-associated signatures for prognostic and immune microenvironment evaluation in hepatocellular carcinoma.Front Immunol. 2022 Aug 23;13:973649. doi: 10.3389/fimmu.2022.973649. eCollection 2022. Front Immunol. 2022. PMID: 36081504 Free PMC article.
-
A necroptosis -related signature for predicting prognosis and immunotherapy in hepatocellular carcinoma.Front Genet. 2022 Sep 5;13:919599. doi: 10.3389/fgene.2022.919599. eCollection 2022. Front Genet. 2022. PMID: 36134033 Free PMC article.
Cited by
-
Identification and characterization of a novel molecular classification based on disulfidptosis-related genes to predict prognosis and immunotherapy efficacy in hepatocellular carcinoma.Aging (Albany NY). 2023 Jul 3;15(13):6135-6151. doi: 10.18632/aging.204809. Epub 2023 Jul 3. Aging (Albany NY). 2023. PMID: 37399661 Free PMC article.
-
Leveraging Diverse Regulated Cell Death Patterns to Identify Diagnosis Biomarkers for Alzheimer's Disease.J Prev Alzheimers Dis. 2024;11(6):1775-1788. doi: 10.14283/jpad.2024.119. J Prev Alzheimers Dis. 2024. PMID: 39559889 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

