Semi-supervised learning in cancer diagnostics
- PMID: 35912249
- PMCID: PMC9329803
- DOI: 10.3389/fonc.2022.960984
Semi-supervised learning in cancer diagnostics
Abstract
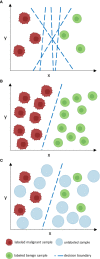
In cancer diagnostics, a considerable amount of data is acquired during routine work-up. Recently, machine learning has been used to build classifiers that are tasked with cancer detection and aid in clinical decision-making. Most of these classifiers are based on supervised learning (SL) that needs time- and cost-intensive manual labeling of samples by medical experts for model training. Semi-supervised learning (SSL), however, works with only a fraction of labeled data by including unlabeled samples for information abstraction and thus can utilize the vast discrepancy between available labeled data and overall available data in cancer diagnostics. In this review, we provide a comprehensive overview of essential functionalities and assumptions of SSL and survey key studies with regard to cancer care differentiating between image-based and non-image-based applications. We highlight current state-of-the-art models in histopathology, radiology and radiotherapy, as well as genomics. Further, we discuss potential pitfalls in SSL study design such as discrepancies in data distributions and comparison to baseline SL models, and point out future directions for SSL in oncology. We believe well-designed SSL models to strongly contribute to computer-guided diagnostics in malignant disease by overcoming current hinderances in the form of sparse labeled and abundant unlabeled data.
Keywords: artificial intelligence; cancer; diagnostics; machine learning; semi-supervised learning.
Copyright © 2022 Eckardt, Bornhäuser, Wendt and Middeke.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- van Engelen JE, Hoos HH. A survey on semi-supervised learning. Mach Learn (2020) 109:373–440. doi: 10.1007/s10994-019-05855-6 - DOI
-
- Triguero I, García S, Herrera F. Self-labeled techniques for semi-supervised learning: taxonomy, software and empirical study. Knowl Inf Syst (2015) 42:245–84. doi: 10.1007/s10115-013-0706-y - DOI
Publication types
LinkOut - more resources
Full Text Sources

