Enteroendocrine cell types that drive food reward and aversion
- PMID: 35913117
- PMCID: PMC9363118
- DOI: 10.7554/eLife.74964
Enteroendocrine cell types that drive food reward and aversion
Abstract
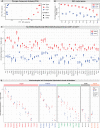
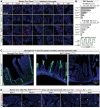
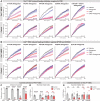
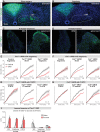
Animals must learn through experience which foods are nutritious and should be consumed, and which are toxic and should be avoided. Enteroendocrine cells (EECs) are the principal chemosensors in the GI tract, but investigation of their role in behavior has been limited by the difficulty of selectively targeting these cells in vivo. Here, we describe an intersectional genetic approach for manipulating EEC subtypes in behaving mice. We show that multiple EEC subtypes inhibit food intake but have different effects on learning. Conditioned flavor preference is driven by release of cholecystokinin whereas conditioned taste aversion is mediated by serotonin and substance P. These positive and negative valence signals are transmitted by vagal and spinal afferents, respectively. These findings establish a cellular basis for how chemosensing in the gut drives learning about food.
Keywords: appetite; feeding; mouse; neuroscience; physiology.
© 2022, Bai et al.
Conflict of interest statement
LB, NS, SY, SM, TD, TL, TC, JG, BJ, ZK No competing interests declared
Figures









References
-
- Borgmann D, Ciglieri E, Biglari N, Brandt C, Cremer AL, Backes H, Tittgemeyer M, Wunderlich FT, Brüning JC, Fenselau H. Gut-brain communication by distinct sensory neurons differently controls feeding and glucose metabolism. Cell Metabolism. 2021;33:1466–1482. doi: 10.1016/j.cmet.2021.05.002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

