The unmitigated profile of COVID-19 infectiousness
- PMID: 35913120
- PMCID: PMC9391043
- DOI: 10.7554/eLife.79134
The unmitigated profile of COVID-19 infectiousness
Abstract
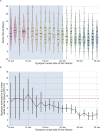
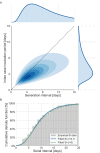
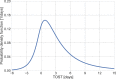
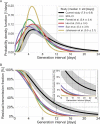
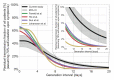
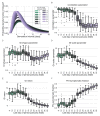
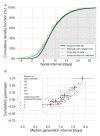
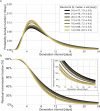
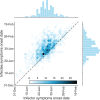
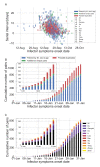
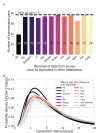
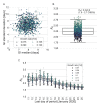
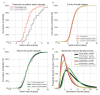
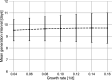
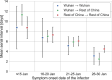
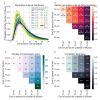
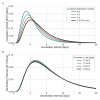
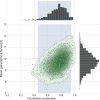
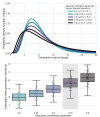
Quantifying the temporal dynamics of infectiousness of individuals infected with SARS-CoV-2 is crucial for understanding the spread of COVID-19 and for evaluating the effectiveness of mitigation strategies. Many studies have estimated the infectiousness profile using observed serial intervals. However, statistical and epidemiological biases could lead to underestimation of the duration of infectiousness. We correct for these biases by curating data from the initial outbreak of the pandemic in China (when mitigation was minimal), and find that the infectiousness profile of the original strain is longer than previously thought. Sensitivity analysis shows our results are robust to model structure, assumed growth rate and potential observational biases. Although unmitigated transmission data is lacking for variants of concern (VOCs), previous analyses suggest that the alpha and delta variants have faster within-host kinetics, which we extrapolate to crude estimates of variant-specific unmitigated generation intervals. Knowing the unmitigated infectiousness profile of infected individuals can inform estimates of the effectiveness of isolation and quarantine measures. The framework presented here can help design better quarantine policies in early stages of future epidemics.
Keywords: COVID-19; SARS-CoV-2; computational biology; epidemiology; generation interval; global health; human; mitigation; systems biology.
© 2022, Sender, Bar-On et al.
Conflict of interest statement
RS, YB, SP, EN, JD, RM No competing interests declared
Figures



Similar articles
-
High infectiousness immediately before COVID-19 symptom onset highlights the importance of continued contact tracing.Elife. 2021 Apr 26;10:e65534. doi: 10.7554/eLife.65534. Elife. 2021. PMID: 33899740 Free PMC article.
-
On realized serial and generation intervals given control measures: The COVID-19 pandemic case.PLoS Comput Biol. 2021 Mar 29;17(3):e1008892. doi: 10.1371/journal.pcbi.1008892. eCollection 2021 Mar. PLoS Comput Biol. 2021. PMID: 33780436 Free PMC article.
-
A Recursive Model of the Spread of COVID-19: Modelling Study.JMIR Public Health Surveill. 2021 Apr 19;7(4):e21468. doi: 10.2196/21468. JMIR Public Health Surveill. 2021. PMID: 33871381 Free PMC article.
-
Pilot Investigation of SARS-CoV-2 Secondary Transmission in Kindergarten Through Grade 12 Schools Implementing Mitigation Strategies - St. Louis County and City of Springfield, Missouri, December 2020.MMWR Morb Mortal Wkly Rep. 2021 Mar 26;70(12):449-455. doi: 10.15585/mmwr.mm7012e4. MMWR Morb Mortal Wkly Rep. 2021. PMID: 33764961 Free PMC article.
-
How to detect and reduce potential sources of biases in studies of SARS-CoV-2 and COVID-19.Eur J Epidemiol. 2021 Feb;36(2):179-196. doi: 10.1007/s10654-021-00727-7. Epub 2021 Feb 25. Eur J Epidemiol. 2021. PMID: 33634345 Free PMC article. Review.
Cited by
-
Disentangling the role of virus infectiousness and awareness-based human behavior during the early phase of the COVID-19 pandemic in the European Union.Appl Math Model. 2023 Oct;122:187-199. doi: 10.1016/j.apm.2023.05.027. Epub 2023 May 29. Appl Math Model. 2023. PMID: 37283821 Free PMC article.
-
Correlation between times to SARS-CoV-2 symptom onset and secondary transmission undermines epidemic control efforts.Epidemics. 2022 Dec;41:100655. doi: 10.1016/j.epidem.2022.100655. Epub 2022 Nov 14. Epidemics. 2022. PMID: 36413921 Free PMC article.
-
Detecting changes in generation and serial intervals under varying pathogen biology, contact patterns and outbreak response.PLoS Comput Biol. 2024 Mar 22;20(3):e1011967. doi: 10.1371/journal.pcbi.1011967. eCollection 2024 Mar. PLoS Comput Biol. 2024. PMID: 38517931 Free PMC article.
-
Inferring the differences in incubation-period and generation-interval distributions of the Delta and Omicron variants of SARS-CoV-2.Proc Natl Acad Sci U S A. 2023 May 30;120(22):e2221887120. doi: 10.1073/pnas.2221887120. Epub 2023 May 22. Proc Natl Acad Sci U S A. 2023. PMID: 37216529 Free PMC article.
-
Best practices for estimating and reporting epidemiological delay distributions of infectious diseases.PLoS Comput Biol. 2024 Oct 28;20(10):e1012520. doi: 10.1371/journal.pcbi.1012520. eCollection 2024 Oct. PLoS Comput Biol. 2024. PMID: 39466727 Free PMC article.
References
-
- Brandal LT, MacDonald E, Veneti L, Ravlo T, Lange H, Naseer U, Feruglio S, Bragstad K, Hungnes O, Ødeskaug LE, Hagen F, Hanch-Hansen KE, Lind A, Watle SV, Taxt AM, Johansen M, Vold L, Aavitsland P, Nygård K, Madslien EH. Outbreak caused by the SARS-cov-2 omicron variant in norway, november to december 2021. Euro Surveillance. 2021;26:2101147. doi: 10.2807/1560-7917.ES.2021.26.50.2101147. - DOI - PMC - PubMed
-
- CDC Science Brief: Options to Reduce Quarantine for Contacts of Persons with SARS-CoV-2 Infection Using Symptom Monitoring and Diagnostic Testing. 2020. [May 9, 2021]. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scienti... - PubMed
-
- CDC CDC Updates and Shortens Recommended Isolation and Quarantine Period for General Population. 2021. [January 2, 2022]. https://www.cdc.gov/media/releases/2021/s1227-isolation-quarantine-guida...
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

