The Integrative Conjugative Element ICESpyM92 Contributes to Pathogenicity of Emergent Antimicrobial-Resistant emm92 Group A Streptococcus
- PMID: 35913172
- PMCID: PMC9387263
- DOI: 10.1128/iai.00080-22
The Integrative Conjugative Element ICESpyM92 Contributes to Pathogenicity of Emergent Antimicrobial-Resistant emm92 Group A Streptococcus
Abstract
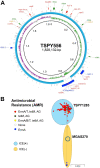
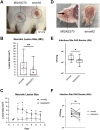
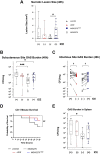
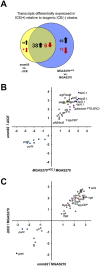
Antimicrobial resistance-encoding mobile genetic elements (MGEs) may contribute to the disease potential of bacterial pathogens. We previously described the association of Group A Streptococcus (GAS) derived from invasive disease with increasingly frequent antimicrobial resistance (AMR). We hypothesized that a 65-kb AMR-encoding MGE (ICESpyM92), highly conserved among closely related emergent invasive emm92 GAS, contributes to GAS disease potential. Here, we provide evidence that a combination of ICESpyM92- and core genome-dependent differential gene expression (DGE) contributes to invasive disease phenotypes of emergent emm92 GAS. Using isogenic ICESpyM92 mutants generated in distinct emm92 genomic backgrounds, we determined the presence of ICESpyM92 enhances GAS virulence in a mouse subcutaneous infection model. Measurement of in vitro and ex vivo DGE indicates ICESpyM92 influences GAS global gene expression in a background-dependent manner. Our study links virulence and AMR on a unique MGE via MGE-related DGE and highlights the importance of investigating associations between AMR-encoding MGEs and pathogenicity.
Keywords: Streptococcus pyogenes; antimicrobial resistance; mobile genetic element; transcriptome; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Virulence Role of the GlcNAc Side Chain of the Lancefield Cell Wall Carbohydrate Antigen in Non-M1-Serotype Group A Streptococcus.mBio. 2018 Jan 30;9(1):e02294-17. doi: 10.1128/mBio.02294-17. mBio. 2018. PMID: 29382733 Free PMC article.
-
Unexpected relationships between frequency of antimicrobial resistance, disease phenotype and emm type in group A Streptococcus.Microb Genom. 2019 Nov;5(11):e000316. doi: 10.1099/mgen.0.000316. Microb Genom. 2019. PMID: 31755853 Free PMC article.
-
Virulence genes, resistome and mobilome of Streptococcus suis strains isolated in France.Microb Genom. 2024 Mar;10(3):001224. doi: 10.1099/mgen.0.001224. Microb Genom. 2024. PMID: 38536216 Free PMC article.
-
Chromosomal islands of Streptococcus pyogenes and related streptococci: molecular switches for survival and virulence.Front Cell Infect Microbiol. 2014 Aug 12;4:109. doi: 10.3389/fcimb.2014.00109. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25161960 Free PMC article. Review.
-
Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions.Trends Microbiol. 2003 May;11(5):224-32. doi: 10.1016/s0966-842x(03)00098-2. Trends Microbiol. 2003. PMID: 12781526 Review.
Cited by
-
Regulation of erm(T) MLSB phenotype expression in the emergent emm92 type group A Streptococcus.NPJ Antimicrob Resist. 2024 Nov 29;2(1):44. doi: 10.1038/s44259-024-00062-3. NPJ Antimicrob Resist. 2024. PMID: 39843607 Free PMC article.
-
LiaR-dependent gene expression contributes to antimicrobial responses in group A Streptococcus.Antimicrob Agents Chemother. 2024 Dec 5;68(12):e0049624. doi: 10.1128/aac.00496-24. Epub 2024 Nov 13. Antimicrob Agents Chemother. 2024. PMID: 39535201 Free PMC article.
-
Identification of a novel CG307 sub-clade in third-generation-cephalosporin-resistant Klebsiella pneumoniae causing invasive infections in the USA.Microb Genom. 2024 Feb;10(2):001201. doi: 10.1099/mgen.0.001201. Microb Genom. 2024. PMID: 38407244 Free PMC article.
-
Emergence of Erythromycin-Resistant Invasive Group A Streptococcus, West Virginia, USA, 2020-2021.Emerg Infect Dis. 2023 May;29(5):898-908. doi: 10.3201/eid2905.221421. Emerg Infect Dis. 2023. PMID: 37080963 Free PMC article.
References
-
- Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu M-Y, Smoot JC, Porcella SF, Parkins LD, Campbell DS, Smith TM, McCormick JK, Leung DYM, Schlievert PM, Musser JM. 2002. Genome sequence of a serotype M3 strain of Group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA 99:10078–10083. 10.1073/pnas.152298499. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

