SDC1-dependent TGM2 determines radiosensitivity in glioblastoma by coordinating EPG5-mediated fusion of autophagosomes with lysosomes
- PMID: 35913916
- PMCID: PMC9980589
- DOI: 10.1080/15548627.2022.2105562
SDC1-dependent TGM2 determines radiosensitivity in glioblastoma by coordinating EPG5-mediated fusion of autophagosomes with lysosomes
Abstract
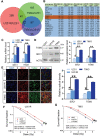
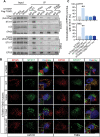
Glioblastoma multiforme (GBM) is the most common brain malignancy insensitive to radiotherapy (RT). Although macroautophagy/autophagy was reported to be a fundamental factor prolonging the survival of tumors under radiotherapeutic stress, the autophagic biomarkers coordinated to radioresistance of GBM are still lacking in clinical practice. Here we established radioresistant GBM cells and identified their protein profiles using tandem mass tag (TMT) quantitative proteomic analysis. It was found that SDC1 and TGM2 proteins were overexpressed in radioresistant GBM cells and tissues and they contributed to the poor prognosis of RT. Knocking down SDC1 and TGM2 inhibited the fusion of autophagosomes with lysosomes and thus enhanced the radiosensitivity of GBM cells. After irradiation, TGM2 bound with SDC1 and transported it from the cell membrane to lysosomes, and then bound to LC3 through its two LC3-interacting regions (LIRs), coordinating the encounter between autophagosomes and lysosomes, which should be a prerequisite for lysosomal EPG5 to recognize LC3 and subsequently stabilize the STX17-SNAP29-VAMP8 QabcR SNARE complex assembly. Moreover, when combined with RT, cystamine dihydrochloride (a TGM2 inhibitor) extended the lifespan of GBM-bearing mice. Overall, our findings demonstrated the EPG5 tethering mode with SDC1 and TGM2 during the fusion of autophagosomes with lysosomes, providing new insights into the molecular mechanism and therapeutic target underlying radioresistant GBM.Abbreviations: BafA1: bafilomycin A1; CQ: chloroquine; Cys-D: cystamine dihydrochloride; EPG5: ectopic P-granules 5 autophagy tethering factor; GBM: glioblastoma multiforme; GFP: green fluorescent protein; LAMP2: lysosomal associated membrane protein 2; LIRs: LC3-interacting regions; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; NC: negative control; RFP: red fluorescent protein; RT: radiotherapy; SDC1: syndecan 1; SNAP29: synaptosome associated protein 29; SQSTM1/p62: sequestosome 1; STX17: syntaxin 17; TGM2: transglutaminase 2; TMT: tandem mass tag; VAMP8: vesicle associated membrane protein 8; WT: wild type.
Keywords: Autophagosome maturation; EPG5; SDC1; TGM2; glioblastoma; radioresistance biomarkers.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
