Genomic surveillance, evolution and global transmission of SARS-CoV-2 during 2019-2022
- PMID: 35913920
- PMCID: PMC9342790
- DOI: 10.1371/journal.pone.0271074
Genomic surveillance, evolution and global transmission of SARS-CoV-2 during 2019-2022
Abstract
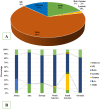
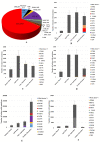
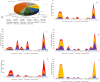
In spite of the availability of vaccine, the health burden associated with the COVID-19 pandemic continues to increase. An estimated 5 million people have died with SARS-CoV-2 infection. Analysis of evolution and genomic diversity can provide sufficient information to reduce the health burden of the pandemic. This study focused to conduct worldwide genomic surveillance. About 7.6 million genomic data were analyzed during 2019 to 2022. Multiple sequence alignment was conducted by using maximum likelihood method. Clade GK (52%) was the most predominant followed by GRY (12%), GRA (11%), GR (8%), GH (7%), G (6%), GV (3%), and O (1%), respectively. VOC Delta (66%) was the most prevalent variant followed by VOC Alpha (18%), VOC Omicron (13%), VOC Gamma (2%) and VOC Beta (1%), respectively. The frequency of point mutations including E484K, N501Y, N439K, and L452R at spike protein has increased 10%-92%. Evolutionary rate of the variants was 23.7 substitution per site per year. Substitution mutations E484K and N501Y had significant correlation with cases (r = .45, r = .23), fatalities (r = .15, r = .44) and growth rate R0 (r = .28, r = .54). This study will help to understand the genomic diversity, evolution and the impact of the variants on the outcome of the COVID-19 pandemic.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

