A complete twelve-gene deletion null mutant reveals that cyclic di-GMP is a global regulator of phase-transition and host colonization in Erwinia amylovora
- PMID: 35914003
- PMCID: PMC9371280
- DOI: 10.1371/journal.ppat.1010737
A complete twelve-gene deletion null mutant reveals that cyclic di-GMP is a global regulator of phase-transition and host colonization in Erwinia amylovora
Abstract
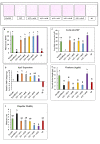
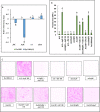
Cyclic-di-GMP (c-di-GMP) is an essential bacterial second messenger that regulates biofilm formation and pathogenicity. To study the global regulatory effect of individual components of the c-di-GMP metabolic system, we deleted all 12 diguanylate cyclase (dgc) and phosphodiesterase (pde)-encoding genes in E. amylovora Ea1189 (Ea1189Δ12). Ea1189Δ12 was impaired in surface attachment due to a transcriptional dysregulation of the type IV pilus and the flagellar filament. A transcriptomic analysis of surface-exposed WT Ea1189 and Ea1189Δ12 cells indicated that genes involved in metabolism, appendage generation and global transcriptional/post-transcriptional regulation were differentially regulated in Ea1189Δ12. Biofilm formation was regulated by all 5 Dgcs, whereas type III secretion and disease development were differentially regulated by specific Dgcs. A comparative transcriptomic analysis of Ea1189Δ8 (lacks all five enzymatically active dgc and 3 pde genes) against Ea1189Δ8 expressing specific dgcs, revealed the presence of a dual modality of spatial and global regulatory frameworks in the c-di-GMP signaling network.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Cyclic Di-GMP modulates the disease progression of Erwinia amylovora.J Bacteriol. 2013 May;195(10):2155-65. doi: 10.1128/JB.02068-12. Epub 2013 Mar 8. J Bacteriol. 2013. PMID: 23475975 Free PMC article.
-
Phosphodiesterase Genes Regulate Amylovoran Production, Biofilm Formation, and Virulence in Erwinia amylovora.Appl Environ Microbiol. 2018 Dec 13;85(1):e02233-18. doi: 10.1128/AEM.02233-18. Print 2019 Jan 1. Appl Environ Microbiol. 2018. PMID: 30366999 Free PMC article.
-
Enzymatically active and inactive phosphodiesterases and diguanylate cyclases are involved in regulation of Motility or sessility in Escherichia coli CFT073.mBio. 2012 Oct 9;3(5):e00307-12. doi: 10.1128/mBio.00307-12. mBio. 2012. PMID: 23047748 Free PMC article.
-
[Activity of cyclic diguanylate (c-di-GMP) in bacteria and the study of its derivatives].Yao Xue Xue Bao. 2012 Mar;47(3):307-12. Yao Xue Xue Bao. 2012. PMID: 22645753 Review. Chinese.
-
Investigating c-di-GMP Signaling in Vibrio parahaemolyticus: Biological Effects and Mechanisms of Regulation.Curr Microbiol. 2025 Jun 2;82(7):319. doi: 10.1007/s00284-025-04260-8. Curr Microbiol. 2025. PMID: 40455311 Review.
Cited by
-
pGpG-signaling regulates virulence and global transcriptomic targets in Erwinia amylovora.bioRxiv [Preprint]. 2024 Jan 14:2024.01.12.575434. doi: 10.1101/2024.01.12.575434. bioRxiv. 2024. PMID: 38260453 Free PMC article. Preprint.
-
Cyclic di-GMP regulates bacterial colonization and further biocontrol efficacy of Bacillus velezensis against apple ring rot disease via its potential receptor YdaK.Front Microbiol. 2022 Dec 16;13:1034168. doi: 10.3389/fmicb.2022.1034168. eCollection 2022. Front Microbiol. 2022. PMID: 36590391 Free PMC article.
-
The global regulation of c-di-GMP and cAMP in bacteria.mLife. 2024 Mar 11;3(1):42-56. doi: 10.1002/mlf2.12104. eCollection 2024 Mar. mLife. 2024. PMID: 38827514 Free PMC article. Review.
-
CsrD regulates amylovoran biosynthesis and virulence in Erwinia amylovora in a novel cyclic-di-GMP dependent manner.Mol Plant Pathol. 2022 Aug;23(8):1154-1169. doi: 10.1111/mpp.13217. Epub 2022 Apr 9. Mol Plant Pathol. 2022. PMID: 35396793 Free PMC article.
References
-
- Solano C, Garcia B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, et al. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci U S A. 2009;106(19):7997–8002. Epub 2009/05/07. doi: 10.1073/pnas.0812573106 ; PubMed Central PMCID: PMC2683120. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

